

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (6): 768-776.DOI: 10.11983/CBB20098 cstr: 32102.14.CBB20098
Previous Articles Next Articles
Yijun Wang1,2,*( ), Yali Wang1,2, Yudong Chen1,2
), Yali Wang1,2, Yudong Chen1,2
Received:2020-05-26
Accepted:2020-09-03
Online:2020-11-01
Published:2020-11-11
Contact:
Yijun Wang
Yijun Wang, Yali Wang, Yudong Chen. Transposon-derived Long Noncoding RNA in Plants[J]. Chinese Bulletin of Botany, 2020, 55(6): 768-776.
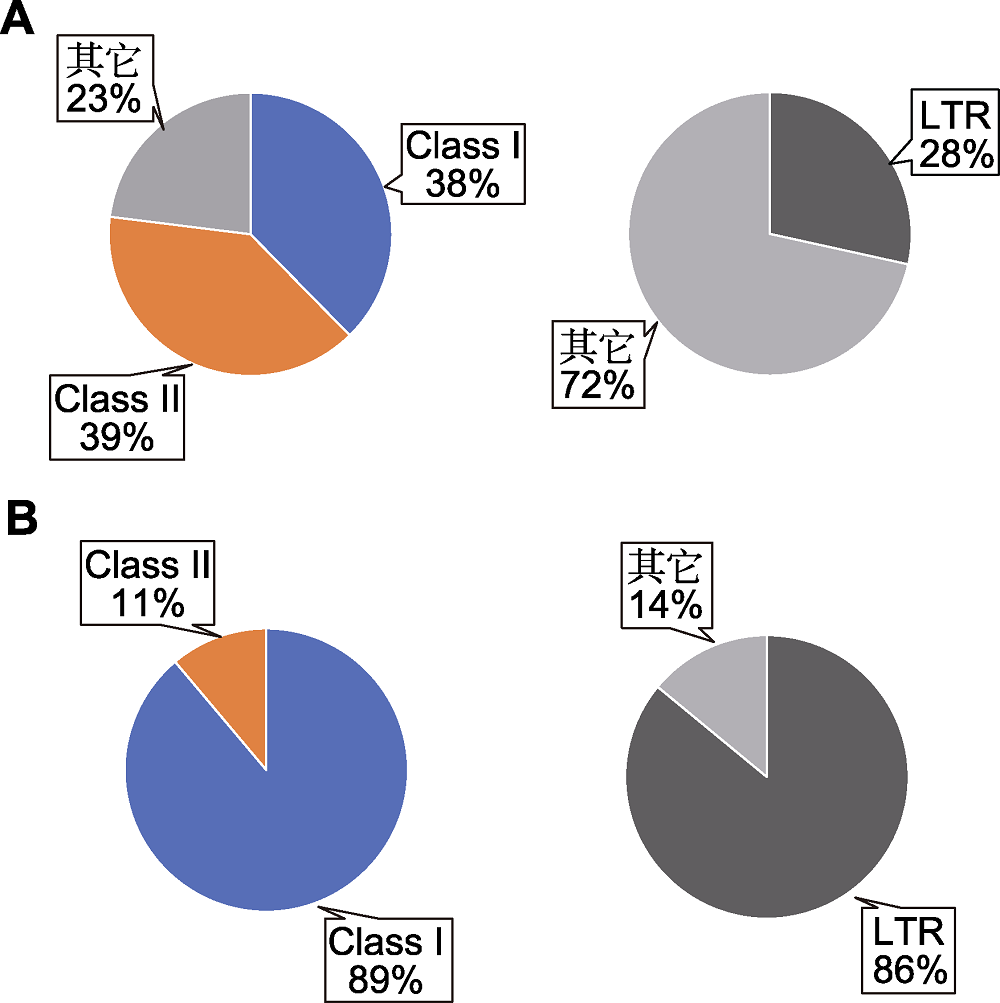
Figure 1 Transposon composition in the genomes of Arabidopsis and maize (data source: The Arabidopsis Genome Initiative, 2000; Schnable et al., 2009) (A) Transposon composition in Arabidopsis genome (left) and the proportion of Arabidopsis long terminal repeat (LTR) retrotransposons (right); (B) Transposon composition in maize genome (left) and the proportion of maize LTR retrotransposons (right). Class I: Retrotransposons; Class II: DNA trotransposons
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
Table 1 Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
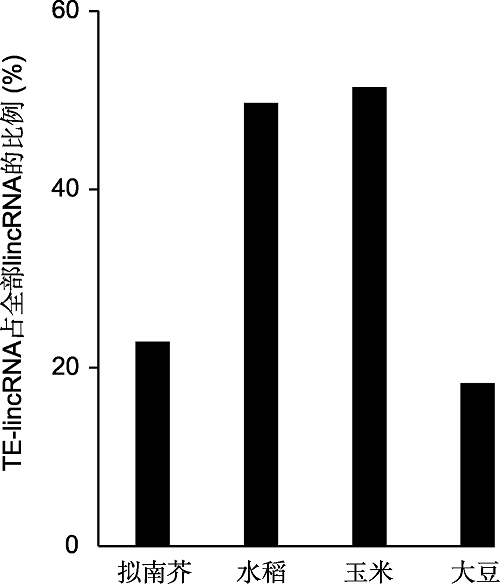
Figure 2 The proportion of plant transposable element-de- rived long intergenic noncoding RNA (TE-lincRNA) (data source: Wang et al., 2017; Golicz et al., 2018)
| [1] | 张楠, 刘自广, 孙世臣, 刘圣怡, 林建辉, 彭疑芳, 张晓旭, 杨贺, 岑曦, 吴娟 (2020). 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用. 植物学报 55, 421-429. |
| [2] | 张硕, 吴昌银 (2019). 长链非编码RNA基因Ef-cd调控水稻早熟与稳产. 植物学报 54, 550-553. |
| [3] |
Alonge M, Wang XG, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, Levy Y, Harel TH, Shalev-Schlosser G, Amsellem Z, Razifard H, Caicedo AL, Tieman DM, Klee H, Kirsche M, Aganezov S, Ranallo-Benavidez TR, Lemmon ZH, Kim J, Robitaille G, Kramer M, Goodwin S, McCombie WR, Hutton S, Van Eck J, Gillis J, Eshed Y, Sedlazeck FJ, van der Knaap E, Schatz MC, Lippman ZB (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145-161.
URL PMID |
| [4] |
Axtell MJ (2013). Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64, 137-159.
URL PMID |
| [5] | Barnes C, Kanhere A(2016). Identification of RNA-protein interactions through in vitro RNA pull-down assays. In: Lanzuolo C, Bodega B, eds. Polycomb Group Proteins. Methods in Molecular Biology, Vol. 480. New York: Humana Press. pp. 99-113. |
| [6] |
Bunkers G, Nelson OE Jr, Raboy V (1993). Maize bronze 1:dSpm insertion mutations that are not fully suppressed by an active Spm Genetics 134, 1211-1220.
URL PMID |
| [7] |
Cong CS, Li YB (2020). Progress on Mutator superfamily. Hereditas 42, 131-144.
DOI URL PMID |
| [8] |
Cui J, Luan YS, Jiang N, Bao H, Meng J (2017). Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J 89, 577-589.
URL PMID |
| [9] |
Deniz Ö, Frost JM, Branco MR (2019). Regulation of transposable elements by DNA modifications. Nat Rev Genet 20, 417-431.
DOI URL PMID |
| [10] |
Dooner HK, Belachew A (1991). Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize. Genetics 129, 855-862.
URL PMID |
| [11] |
Du QG, Wang K, Zou C, Xu C, Li WX (2018). The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol 177, 1743-1753.
URL PMID |
| [12] |
Fang J, Zhang FT, Wang HR, Wang W, Zhao F, Li ZJ, Sun CH, Chen FM, Xu F, Chang SQ, Wu L, Bu QY, Wang PR, Xie JK, Chen F, Huang XH, Zhang YJ, Zhu XG, Han B, Deng XJ, Chu CC (2019). Ef-cd locus shortens rice maturity duration without yield penalty. Proc Natl Acad Sci USA 116, 18717-18722.
URL PMID |
| [13] |
Feschotte C, Jiang N, Wessler SR (2002). Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3, 329-341.
DOI URL PMID |
| [14] | Gagliardi M, Matarazzo MR (2016). RIP: RNA immunoprecipitation. In: Lanzuolo C, Bodega B, eds. Polycomb Group Proteins. Methods in Molecular Biology, Vol. 1480. New York: Humana Press.pp. 73-86. |
| [15] |
Golicz AA, Singh MB, Bhalla PL (2018). The long intergenic noncoding RNA (lincRNA) landscape of the soybean genome. Plant Physiol 176, 2133-2147.
URL PMID |
| [16] |
Heo JB, Sung S (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76-79.
DOI URL |
| [17] |
Hou XX, Cui J, Liu WW, Jiang N, Zhou XX, Qi HY, Meng J, Luan YS (2020). LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression. Phytopathology 110, 873-880.
DOI URL PMID |
| [18] |
Huang L, Dong H, Zhou D, Li M, Liu YH, Zhang F, Feng YY, Yu DL, Lin S, Cao JS (2018). Systematic identification of long non-coding RNAs during pollen development and fertilization inBrassica rapa. Plant J 96, 203-222.
DOI URL PMID |
| [19] |
Kapitonov VV, Jurka J (2001). Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA 98, 8714-8719.
DOI URL PMID |
| [20] |
Khanduja JS, Calvo IA, Joh RI, Hill IT, Motamedi M (2016). Nuclear noncoding RNAs and genome stability. Mol Cell 63, 7-20.
DOI URL PMID |
| [21] |
Kidwell MG, Lisch DR (2000). Transposable elements and host genome evolution. Trends Ecol Evol 15, 95-99.
DOI URL PMID |
| [22] |
Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, Evans MMS, Scanlon MJ, Yu JM, Schnable PS, Timmermans MCP, Springer NM, Muehlbauer GJ (2014). Genome- wide discovery and characterization of maize long non- coding RNAs. Genome Biol 15, R40.
DOI URL PMID |
| [23] |
Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1, E67.
DOI URL PMID |
| [24] |
Liu F, Xu YR, Chang KX, Li S, Liu ZG, Qi SD, Jia JB, Zhang M, Crawford NM, Wang Y (2019). The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. New Phytol 224, 117-131.
URL PMID |
| [25] |
Liu HJ, Jian LM, Xu JT, Zhang QH, Zhang ML, Jin ML, Peng Y, Yan JL, Han BZ, Liu J, Gao F, Liu XG, Huang L, Wei WJ, Ding YX, Yang XF, Li ZX, Zhang ML, Sun JM, Bai MJ, Song WH, Chen HM, Sun XA, Li WQ, Lu YM, Liu Y, Zhao JR, Qian YW, Jackson D, Fernie AR, Yan JB (2020). High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 32, 1397-1413.
DOI URL PMID |
| [26] |
Liu J, Jung C, Xu J, Wang H, Deng SL, Bernad L, Arenas-Huertero C, Chua NH (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333-4345.
DOI URL PMID |
| [27] |
Liu X, Li DY, Zhang DL, Yin DD, Zhao Y, Ji CJ, Zhao XF, Li XB, He Q, Chen RS, Hu SN, Zhu LH (2018). A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol 218, 774-788.
DOI URL PMID |
| [28] |
Lv YD, Hu FQ, Zhou YF, Wu FL, Gaut BS (2019). Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response. BMC Genomics 20, 864.
DOI URL PMID |
| [29] |
Martin SL, Garfinkel DJ (2003). Survival strategies for transposons and genomes. Genome Biol 4, 313.
DOI URL PMID |
| [30] |
Qin T, Zhao HY, Cui P, Albesher N, Xiong LM (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 175, 1321-1336.
DOI URL PMID |
| [31] |
Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du FY, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen WZ, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He RF, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin JK, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan CZ, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren LY, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han YJ, Lee H, Li PH, Lisch DR, Liu SZ, Liu ZJ, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang LX, Yu Y, Zhang LF, Zhou SG, Zhu QH, Bennetzen JL, Dawe RK, Jiang JM, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112-1115.
URL PMID |
| [32] |
Seo JS, Sun HX, Park BS, Huang CH, Yeh SD, Jung C, Chua NH (2017). ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29, 1024-1038.
URL PMID |
| [33] |
Swiezewski S, Liu FQ, Magusin A, Dean C (2009). Cold- induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462, 799-802.
DOI URL PMID |
| [34] |
The Arabidopsis Genome Initiative(2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796-815.
DOI URL PMID |
| [35] |
Ulitsky I (2018). Interactions between short and long noncoding RNAs. FEBS Lett 592, 2874-2883.
URL PMID |
| [36] |
Wang D, Qu ZP, Yang L, Zhang QZ, Liu ZH, Do T, Adelson DL, Wang ZY, Searle I, Zhu JK (2017). Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J 90, 133-146.
URL PMID |
| [37] |
Wang X, Ai G, Zhang CL, Cui L, Wang JF, Li HX, Zhang JH, Ye ZB (2016). Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytol 209, 1442-1455.
DOI URL PMID |
| [38] |
Wang Y, Luo XJ, Sun F, Hu JH, Zha XJ, Su W, Yang JS (2018a). aOverexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat Commun 9, 3516.
DOI URL PMID |
| [39] |
Wang YC, Xu JY, Ge M, Ning LH, Hu MM, Zhao H (2020). High-resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize. BMC Genomics 21, 353.
DOI URL PMID |
| [40] |
Wang YJ, Wang YL, Zhao J, Huang JY, Shi YN, Deng DX (2018b). Unveiling gibberellin-responsive coding and long noncoding RNAs in maize. Plant Mol Biol 98, 427-438.
DOI URL PMID |
| [41] |
Wu H, Yang L, Chen LL (2017). The diversity of long noncoding RNAs and their generation. Trends Genet 33, 540-552.
URL PMID |
| [42] |
Xu JH, Messing J (2006). Maize haplotype with a helitron- amplified cytidine deaminase gene copy. BMC Genet 7, 52.
DOI URL PMID |
| [43] |
Yang DW, Li SP, Cui HT, Zou SH, Wang W (2020). Molecular genetic mechanisms of interaction between host plants and pathogens. Hereditas 42, 278-286.
DOI URL PMID |
| [44] |
Yoon JH, Abdelmohsen K, Gorospe M (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol 34, 9-14.
DOI URL PMID |
| [45] |
Yu Y, Zhou YF, Feng YZ, He H, Lian JP, Yang YW, Lei MQ, Zhang YC, Chen YQ (2020). Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol J 18, 679-690.
DOI URL PMID |
| [46] |
Zhao SR, Zhang Y, Gordon W, Quan J, Xi HL, Du S, von Schack D, Zhang BH (2015). Comparison of stranded and non-stranded RNA-seq transcriptome profiling and investigation of gene overlap. BMC Genomics 16, 675.
DOI URL PMID |
| [47] |
Zhao T, Tao XY, Feng SL, Wang LY, Hong H, Ma W, Shang GD, Guo SS, He YX, Zhou BL, Guan XY (2018a). LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol 19, 195.
DOI URL PMID |
| [48] |
Zhao XY, Li JR, Lian B, Gu HQ, Li Y, Qi YJ (2018b). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun 9, 5056.
DOI URL PMID |
| [49] |
Zheng XM, Chen J, Pang HB, Liu S, Gao Q, Wang JR, Qiao WH, Wang H, Liu J, Olsen KM, Yang QW (2019). Genome-wide analyses reveal the role of noncoding variation in complex traits during rice domestication. Sci Adv 5, eaax3619.
DOI URL PMID |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||