

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (6): 893-904.DOI: 10.11983/CBB22231 cstr: 32102.14.CBB22231
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Wenjing Wang1,†, Yu Zhu1,†, Hongming Zhang1,2, Ludan Wei1, Qingping Yi3, Xiaomin Yu1, Yuhan Liu1, Lixue Zhang1, Wenhan Cheng3, Yanhong He1,*( )
)
Received:2022-10-03
Accepted:2023-03-07
Online:2023-11-01
Published:2023-11-27
Contact:
* E-mail: hyh2010@mail.hzau.edu.cn
About author:† These authors contributed equally to this paper
Wenjing Wang, Yu Zhu, Hongming Zhang, Ludan Wei, Qingping Yi, Xiaomin Yu, Yuhan Liu, Lixue Zhang, Wenhan Cheng, Yanhong He. Morphological Identification and Development of Linkage Markers for Lobed Ray Floret Mutants in Marigold (Tagetes erecta)[J]. Chinese Bulletin of Botany, 2023, 58(6): 893-904.
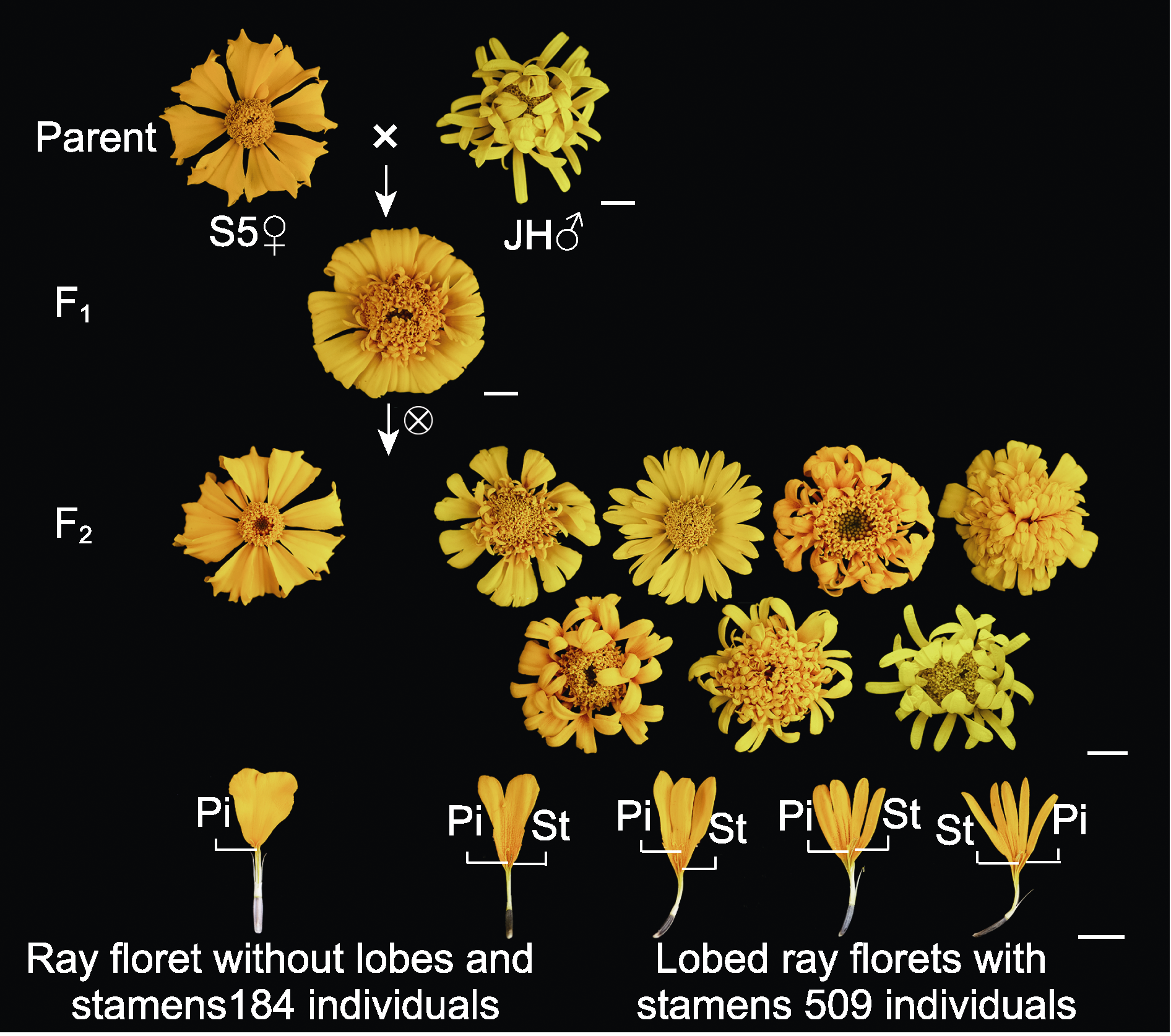
Figure 1 Construction of F2 population between marigold S5 and JH, and segregation of ray corolla traits in the F2 population Pi: Pistil; St: Stamen. Bars=1 cm
| Unigene ID | Primer name | Forward primer (5'-3') | Reverse primer (5'-3') | Restriction enzyme |
|---|---|---|---|---|
| T01.PB44152 | 44152-CAPS | GTCAGCATGGGAATTCTACTGTTGG | CCATTTATAGCTGGAAAACAACCCG | DraI |
| T01.PB49370 | 49370-CAPS | TCGTCTCCACCATAACCACCGC | CTCAACTTTATCCGCGCCAATA | XhoI |
| T01.PB23497 | 23497-CAPS | TTCGTCCAAACCCAGATATGCTTAT | ACCAACCGACAAGTACGATTCGTAT | XbaI |
| T01.PB40376 | 40376-CAPS | GTGTGCGTCTCTGGCACCATCAT | TAAGTACCATTCTCCGACCCGCA | DraI |
| T01.PB44341 | 44341-CAPS | CCAACCTTTTTGTGCTTGCTAAC | AAGTTCCAAGTCTGTTACTTCCATC | XbaI |
| T01.PB36031 | 36031-CAPS | CGGTGACATTATGAACCTTGGACTG | CCCATAACAACAACTTAGTACCGTATT | EcoRI |
| T01.PB21624 | 21624-CAPS | CCGAGCATATTGAATTTTGGGTTAT | AGGGAATGAGAAGAACAATGGTCGT | SalI |
| T01.PB37003 | 37003-CAPS | TGACTCTTTCTATCGCAGAGCCACT | ATCCAGGTACAAGTCACATGTCTGT | EcoRI |
| T01.PB34032 | 34032-CAPS | GAGGTCTGCTTGCTTTTATGAGTAC | GCAAATATGGGCTACAAAATTCTATT | EcoRI |
| T01.PB26228 | 26228-CAPS | AATAGTCCAGCAGCAAAAGTTGGTC | GCTTTCAGTGAACCAAAGGATATGG | XbaI |
| T01.PB17555 | 17555-CAPS | TGCTCATACATCCATCTCCGTCTTC | CAGCTTTCTTGAACGTGTCCCTAAC | XbaI |
| T01.PB43155 | 43155-CAPS | GTATATGCTGCAACTCTGCCCAAAG | GGAACACGATCCATGTCAGGTG | EcoRI |
| T01.PB17261 | 17261-CAPS | GAGAAATACGAACCGTTATGG | AACTAGTACAACATTTTAAACCTGG | EcoRI |
| T01.PB40072 | 40072-CAPS | ATGCAGATGACCCTACAAATTACCT | GCCTCTATTCCTATTTTCTCCTACC | SalI |
Table 1 Primers for amplified polymorphic sequences (CAPS) markers
| Unigene ID | Primer name | Forward primer (5'-3') | Reverse primer (5'-3') | Restriction enzyme |
|---|---|---|---|---|
| T01.PB44152 | 44152-CAPS | GTCAGCATGGGAATTCTACTGTTGG | CCATTTATAGCTGGAAAACAACCCG | DraI |
| T01.PB49370 | 49370-CAPS | TCGTCTCCACCATAACCACCGC | CTCAACTTTATCCGCGCCAATA | XhoI |
| T01.PB23497 | 23497-CAPS | TTCGTCCAAACCCAGATATGCTTAT | ACCAACCGACAAGTACGATTCGTAT | XbaI |
| T01.PB40376 | 40376-CAPS | GTGTGCGTCTCTGGCACCATCAT | TAAGTACCATTCTCCGACCCGCA | DraI |
| T01.PB44341 | 44341-CAPS | CCAACCTTTTTGTGCTTGCTAAC | AAGTTCCAAGTCTGTTACTTCCATC | XbaI |
| T01.PB36031 | 36031-CAPS | CGGTGACATTATGAACCTTGGACTG | CCCATAACAACAACTTAGTACCGTATT | EcoRI |
| T01.PB21624 | 21624-CAPS | CCGAGCATATTGAATTTTGGGTTAT | AGGGAATGAGAAGAACAATGGTCGT | SalI |
| T01.PB37003 | 37003-CAPS | TGACTCTTTCTATCGCAGAGCCACT | ATCCAGGTACAAGTCACATGTCTGT | EcoRI |
| T01.PB34032 | 34032-CAPS | GAGGTCTGCTTGCTTTTATGAGTAC | GCAAATATGGGCTACAAAATTCTATT | EcoRI |
| T01.PB26228 | 26228-CAPS | AATAGTCCAGCAGCAAAAGTTGGTC | GCTTTCAGTGAACCAAAGGATATGG | XbaI |
| T01.PB17555 | 17555-CAPS | TGCTCATACATCCATCTCCGTCTTC | CAGCTTTCTTGAACGTGTCCCTAAC | XbaI |
| T01.PB43155 | 43155-CAPS | GTATATGCTGCAACTCTGCCCAAAG | GGAACACGATCCATGTCAGGTG | EcoRI |
| T01.PB17261 | 17261-CAPS | GAGAAATACGAACCGTTATGG | AACTAGTACAACATTTTAAACCTGG | EcoRI |
| T01.PB40072 | 40072-CAPS | ATGCAGATGACCCTACAAATTACCT | GCCTCTATTCCTATTTTCTCCTACC | SalI |
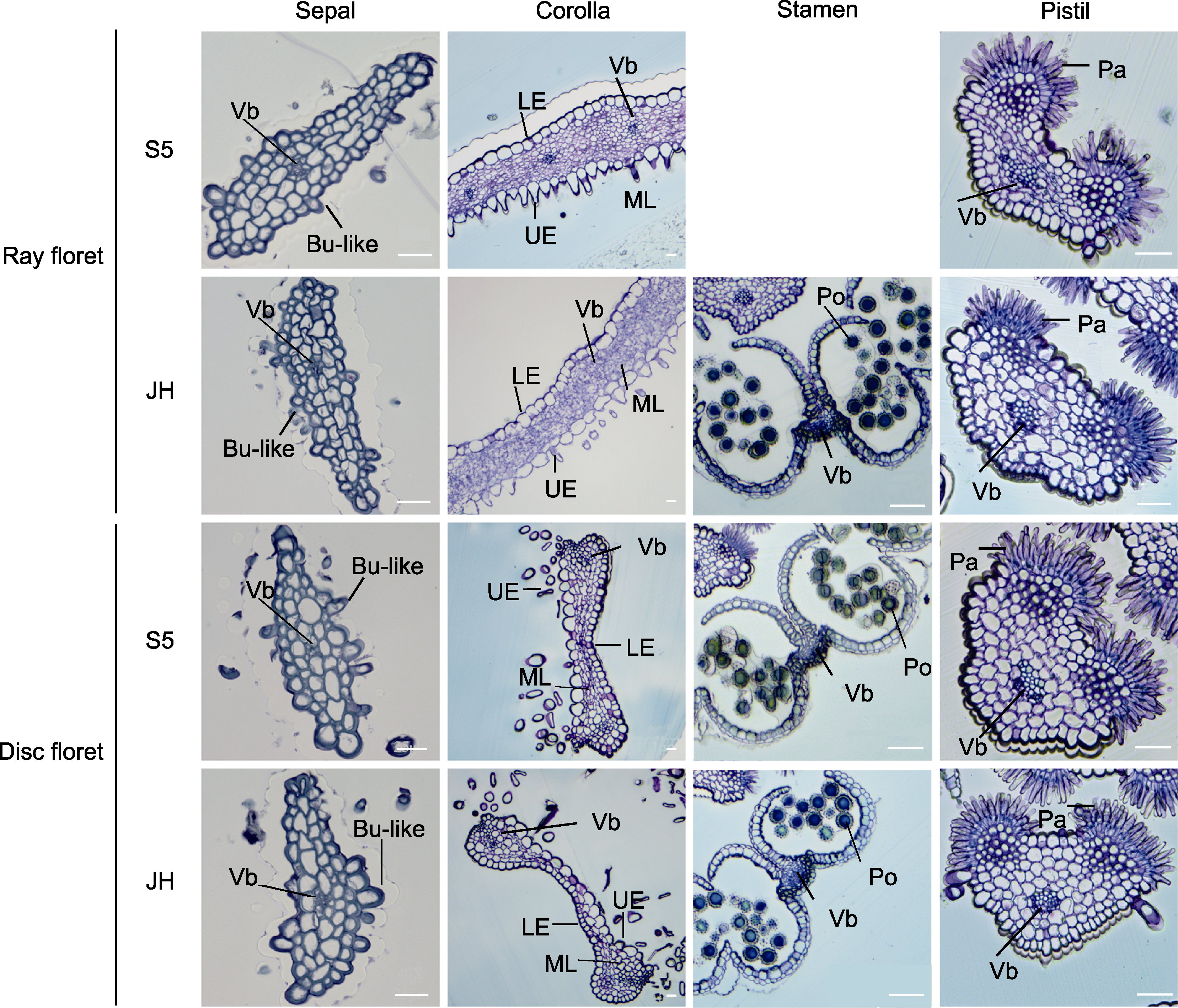
Figure 3 The observation of transverse semi-thin sections in four whorls floral organs of marigold S5 and JH at full blooming stage Vb: Vascular bundles; Bu-like: Bubble-like cells; UE: Upper epidermis; ML: Middle layer; LE: Lower epidermis; Po: Pollen grains; Pa: Papilla cells. Bars=25 μm
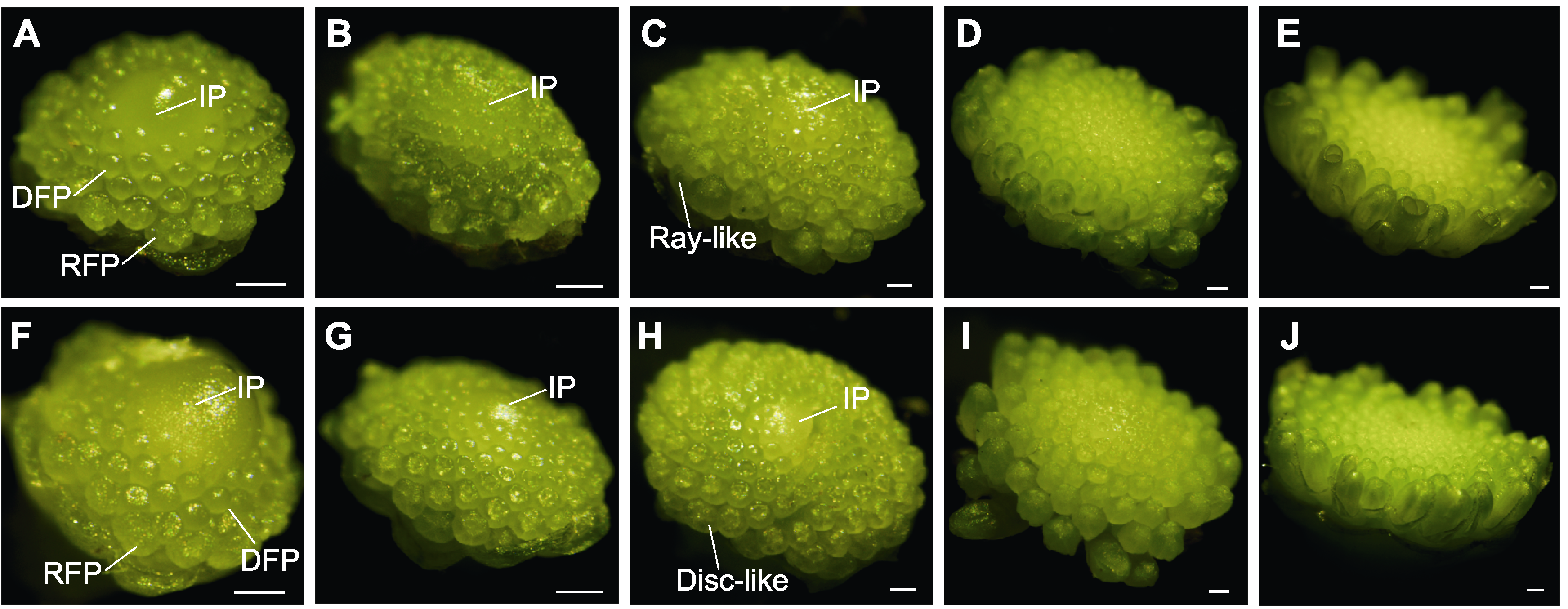
Figure 4 Stereomicroscope observation of floral buds of marigold S5 (A-E) and JH (F-J) at different developmental stages (A), (F) 1 mm buds; (B), (G) 1.5 mm buds; (C), (H) 2 mm buds; (D), (I) 3 mm buds; (E), (J) 4 mm buds. IP: Inflorescence primordium; RFP: Ray floret primordium; DFP: Disc floret primordium; Disc-like: Disc-like floret corolla; Ray-like: Ray-like floret corolla. Bars=200 μm
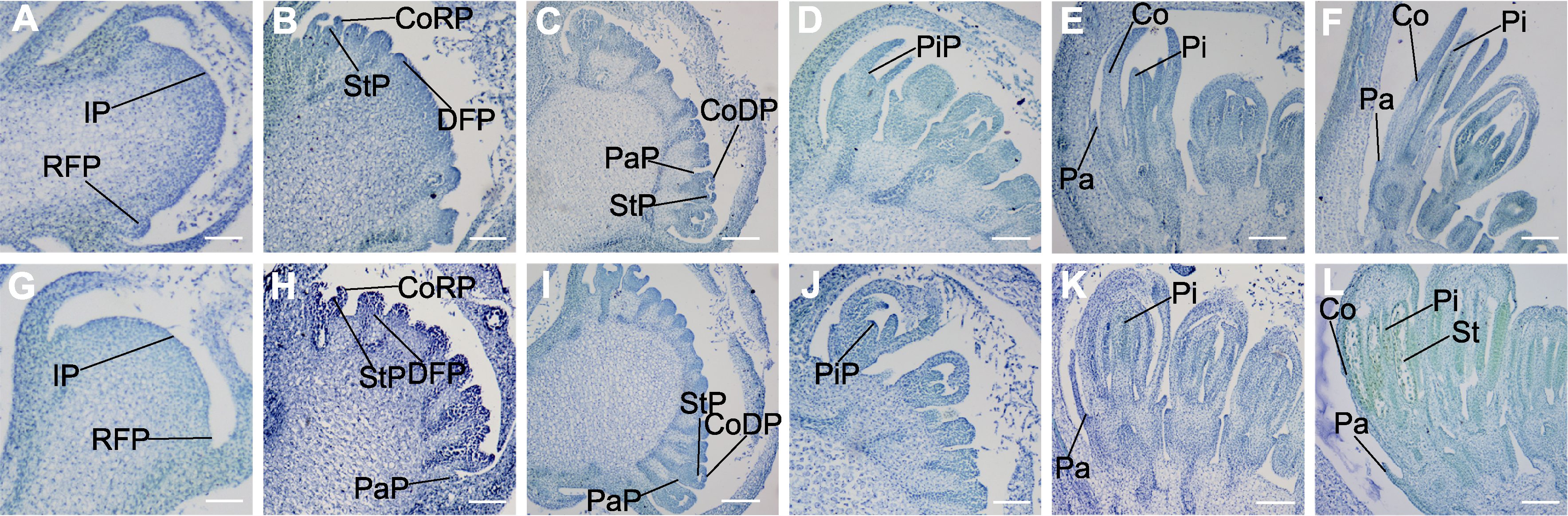
Figure 5 The transverse section of paraffin sections of floral buds of marigold S5 (A-F) and JH (G-L) at different developmental stages (A), (G) 0.5 mm buds; (B), (H) 1 mm buds; (C), (I) 1.5 mm buds; (D), (J) 2 mm buds; (E), (K) 3 mm buds; (F), (L) 4 mm buds. IP: Inflorescence primordium; RFP: Ray floret primordium; DFP: Disc floret primordium; PaP: Pappus primordium; CoRP: Corolla primordium of ray floret; CoDP: Corolla primordium of disc floret; StP: Stamen primordium; PiP: Pistil primordium; Pa: Pappus; Co: Corolla; St: Stamen; Pi: Pistil. Bars=100 μm
| S5-pool | JH-pool | F2-S5 | F2-JH | |
|---|---|---|---|---|
| Total reads | 75274490 | 93514368 | 68385858 | 74073254 |
| Clean reads | 72943532 | 90821192 | 66109078 | 71708520 |
| Total base | 11291173500 | 14027155200 | 10257878700 | 11110988100 |
| Clean base | 10838757206 | 13499502354 | 9820452668 | 10648842550 |
| GC content (%) | 42 | 43 | 43 | 43 |
| Q20 (%) | 99.99 | 100 | 100 | 100 |
| Q30 (%) | 99.57 | 99.63 | 99.47 | 99.55 |
Table 2 Quality of BSR-seq
| S5-pool | JH-pool | F2-S5 | F2-JH | |
|---|---|---|---|---|
| Total reads | 75274490 | 93514368 | 68385858 | 74073254 |
| Clean reads | 72943532 | 90821192 | 66109078 | 71708520 |
| Total base | 11291173500 | 14027155200 | 10257878700 | 11110988100 |
| Clean base | 10838757206 | 13499502354 | 9820452668 | 10648842550 |
| GC content (%) | 42 | 43 | 43 | 43 |
| Q20 (%) | 99.99 | 100 | 100 | 100 |
| Q30 (%) | 99.57 | 99.63 | 99.47 | 99.55 |
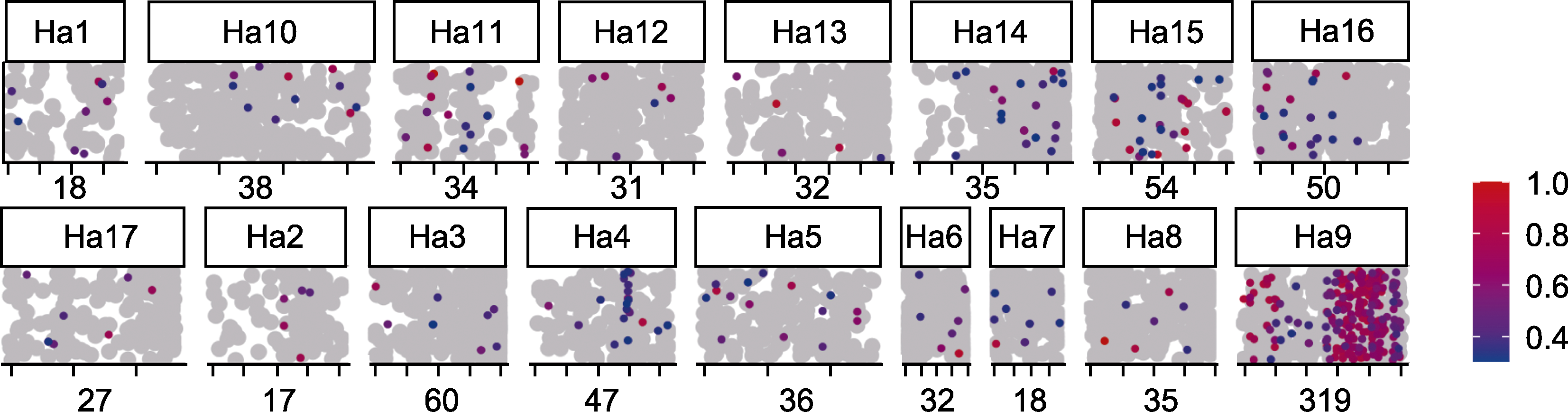
Figure 6 Scatter diagram of homologous gene alignment between marigold transcriptome and sunflower genome based on the results of BSR-seq analysis Grey dots: Locations of homologous genes in marigold mapped to sunflower; Colorful dots: Unigenes in marigold with ΔSNP index≥0.3
| Unigene ID | Primer name | Position | ΔSNP index | Mutant position | Mutant base |
|---|---|---|---|---|---|
| T01.PB44152 | 44152-CAPS | 32539231 | 0.873 | 2113 | A→T |
| T01.PB49370 | 49370-CAPS | 50400088 | 0.763 | 144 | G→T |
| T01.PB23497 | 23497-CAPS | 138318439 | 0.555 | 975 | A→G |
| T01.PB40376 | 40376-CAPS | 158179471 | 0.751 | 402 | T→A |
| T01.PB44341 | 44341-CAPS | 174818902 | 0.666 | 225 | T→C |
| T01.PB36031 | 36031-CAPS | 176617357 | 0.715 | 1712 | A→G |
| T01.PB21624 | 21624-CAPS | 189237745 | 0.630 | 220 | G→C |
| T01.PB37003 | 37003-SCAR | 191264434 | 0.711 | 1054 | T→A |
| T01.PB34032 | 34032-CAPS | 199916490 | 0.846 | 1138 | G→A |
| T01.PB26228 | 26228-CAPS | 204195845 | 0.533 | 1471 | C→T |
| T01.PB17555 | 17555-CAPS | 208494177 | 0.740 | 343 | C→T |
| T01.PB43155 | 43155-CAPS | 210482932 | 0.653 | 1475 | T→C |
| T01.PB17261 | 17261-CAPS | 238981210 | 0.553 | 1316 | A→C |
| T01.PB40072 | 40072-CAPS | 250623369 | 0.555 | 2109 | C→T |
Table 3 The SNPs associated with lobes of ray florets in marigold
| Unigene ID | Primer name | Position | ΔSNP index | Mutant position | Mutant base |
|---|---|---|---|---|---|
| T01.PB44152 | 44152-CAPS | 32539231 | 0.873 | 2113 | A→T |
| T01.PB49370 | 49370-CAPS | 50400088 | 0.763 | 144 | G→T |
| T01.PB23497 | 23497-CAPS | 138318439 | 0.555 | 975 | A→G |
| T01.PB40376 | 40376-CAPS | 158179471 | 0.751 | 402 | T→A |
| T01.PB44341 | 44341-CAPS | 174818902 | 0.666 | 225 | T→C |
| T01.PB36031 | 36031-CAPS | 176617357 | 0.715 | 1712 | A→G |
| T01.PB21624 | 21624-CAPS | 189237745 | 0.630 | 220 | G→C |
| T01.PB37003 | 37003-SCAR | 191264434 | 0.711 | 1054 | T→A |
| T01.PB34032 | 34032-CAPS | 199916490 | 0.846 | 1138 | G→A |
| T01.PB26228 | 26228-CAPS | 204195845 | 0.533 | 1471 | C→T |
| T01.PB17555 | 17555-CAPS | 208494177 | 0.740 | 343 | C→T |
| T01.PB43155 | 43155-CAPS | 210482932 | 0.653 | 1475 | T→C |
| T01.PB17261 | 17261-CAPS | 238981210 | 0.553 | 1316 | A→C |
| T01.PB40072 | 40072-CAPS | 250623369 | 0.555 | 2109 | C→T |
| CAPS markers | F2-S5 | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| 36031-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| 21624-CAPS | * | |||||||||||||||||||||||||||||
| 37003-SCAR | * | |||||||||||||||||||||||||||||
| 34032-CAPS | * | * | ||||||||||||||||||||||||||||
| 44152-CAPS | * | * | * | |||||||||||||||||||||||||||
| 49370-CAPS | * | * | * | |||||||||||||||||||||||||||
| 17555-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| 43155-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| CAPS markers | F2-JH | |||||||||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| 36031-CAPS | * | * | ||||||||||||||||||||||||||||
| 21624-CAPS | * | |||||||||||||||||||||||||||||
| 37003-SCAR | ||||||||||||||||||||||||||||||
| 34032-CAPS | * | |||||||||||||||||||||||||||||
| 44152-CAPS | * | |||||||||||||||||||||||||||||
| 49370-CAPS | * | |||||||||||||||||||||||||||||
| 17555-CAPS | * | * | ||||||||||||||||||||||||||||
| 43155-CAPS | * | * | ||||||||||||||||||||||||||||
Table 4 The individuals with the exchange of molecular markers for eight genes in marigold
| CAPS markers | F2-S5 | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| 36031-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| 21624-CAPS | * | |||||||||||||||||||||||||||||
| 37003-SCAR | * | |||||||||||||||||||||||||||||
| 34032-CAPS | * | * | ||||||||||||||||||||||||||||
| 44152-CAPS | * | * | * | |||||||||||||||||||||||||||
| 49370-CAPS | * | * | * | |||||||||||||||||||||||||||
| 17555-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| 43155-CAPS | * | * | * | * | ||||||||||||||||||||||||||
| CAPS markers | F2-JH | |||||||||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| 36031-CAPS | * | * | ||||||||||||||||||||||||||||
| 21624-CAPS | * | |||||||||||||||||||||||||||||
| 37003-SCAR | ||||||||||||||||||||||||||||||
| 34032-CAPS | * | |||||||||||||||||||||||||||||
| 44152-CAPS | * | |||||||||||||||||||||||||||||
| 49370-CAPS | * | |||||||||||||||||||||||||||||
| 17555-CAPS | * | * | ||||||||||||||||||||||||||||
| 43155-CAPS | * | * | ||||||||||||||||||||||||||||
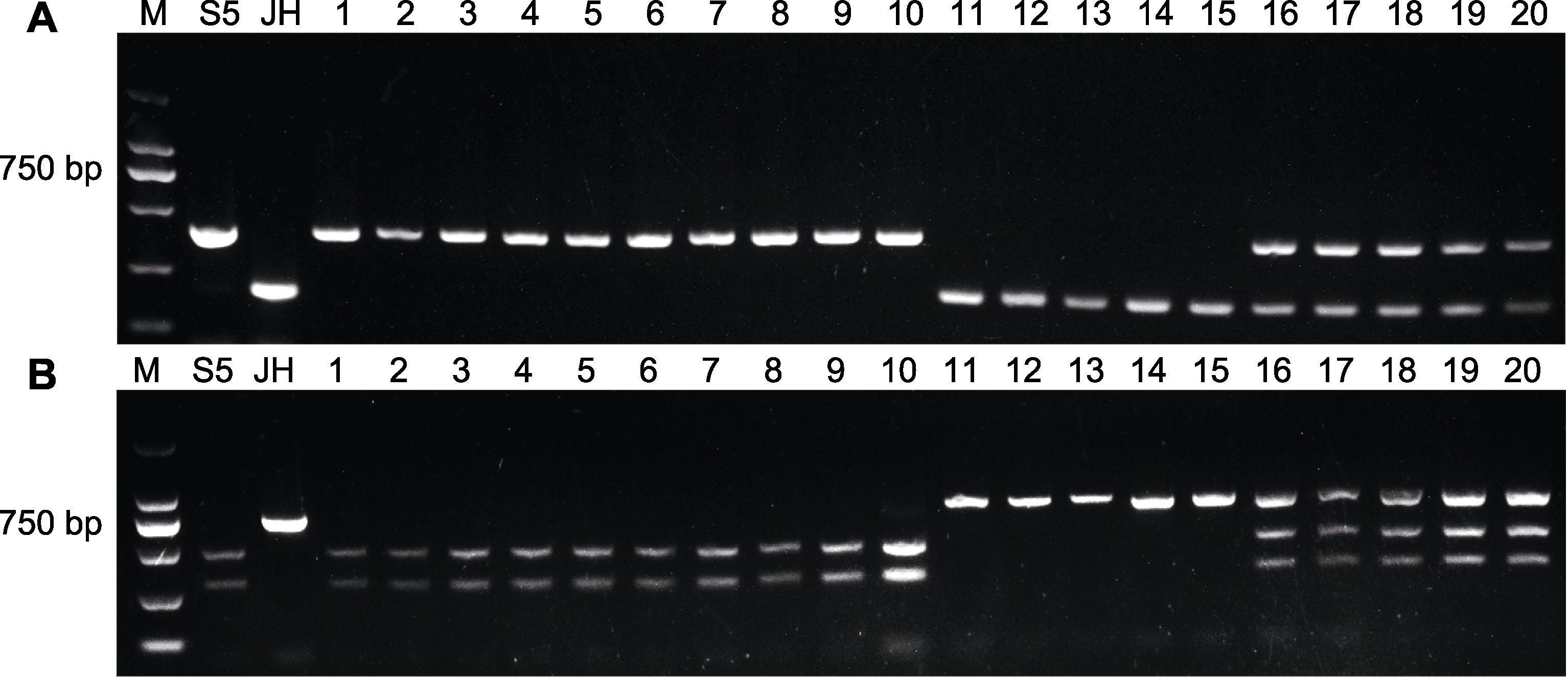
Figure 7 PCR amplification results of 37003-SCAR marker (A) and 34032-CAPS marker (B) in parents and F2 population M: 2 000 bp marker; 1-10: Individuals of F2 with ray floret without lobes and stamens; 11-20: Individuals of F2 that have ray florets with five-lobed and stamens, of which 11-15 are homozygotes and 16-20 are heterozygotes.
| [1] | 陈笛, 刘家亮, 孟祥春, 王小菁 (2006). 南美蟛蜞菊花的生长发育. 植物学通报 23, 37-43. |
| [2] | 何燕红 (2010). 万寿菊雄性不育性状的遗传分析及其育种应用. 博士论文. 武汉: 华中农业大学. pp. 24-25. |
| [3] | 夏伟康 (2020). 菊花转录因子CmCUC2和CmCUC3的功能分析. 硕士论文. 南京: 南京农业大学. pp. 50-51. |
| [4] |
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215, 403-410.
DOI PMID |
| [5] |
Bar M, Ori N (2014). Leaf development and morphogenesis. Development 141, 4219-4230.
DOI PMID |
| [6] |
Bar M, Ori N (2015). Compound leaf development in model plant species. Curr Opin Plant Biol 23, 61-69.
DOI PMID |
| [7] |
Bello MA, Álvarez I, Torices R, Fuertes-Aguilar J (2013). Floral development and evolution of capitulum structure in Anacyclus (Anthemideae, Asteraceae). Ann Bot 112, 1597-1612.
DOI URL |
| [8] |
Blein T, Hasson A, Laufs P (2010). Leaf development: what it needs to be complex. Curr Opin Plant Biol 13, 75-82.
DOI PMID |
| [9] |
Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
DOI PMID |
| [10] |
Broholm SK, Täehtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105, 9117-9122.
DOI PMID |
| [11] |
Chapman MA, Tang SX, Draeger D, Nambeesan S, Shaffer H, Barb JG, Knapp SJ, Burke JM (2012). Genetic analysis of floral symmetry in Van Gogh's sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet 8, e1002628.
DOI URL |
| [12] |
Chen J, Shen CZ, Guo YP, Rao GY (2018). Patterning the Asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like genes. Front Plant Sci 9, 551.
DOI PMID |
| [13] |
Cheng PL, Liu YN, Yang YM, Chen H, Cheng H, Hu Q, Zhang ZX, Gao JJ, Zhang JX, Ding L, Fang WM, Chen SM, Chen FD, Jiang JF (2020). CmBES1 is a regulator of boundary formation in chrysanthemum ray florets. Hortic Res 7, 129.
DOI |
| [14] |
Dadpour MR, Naghiloo S, Gohari G (2011). Inflorescence and floral ontogeny in Osteospermum ecklonis (Asteraceae). Botany 89, 605-614.
DOI URL |
| [15] |
Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D (2004). The role of JAGGED in shaping lateral organs. Development 131, 1101-1110.
DOI PMID |
| [16] |
Endress PK, Matthews ML (2006). Elaborate petals and staminodes in eudicots: diversity, function, and evolution. Org Divers Evol 6, 257-293.
DOI URL |
| [17] |
Fambrini M, Salvini M, Pugliesi C (2011). A transposon-mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus annuus. Genetica 139, 1521-1529.
DOI PMID |
| [18] |
Fu XH, Shan HY, Yao X, Cheng J, Jiang YC, Yin XF, Kong HZ (2022). Petal development and elaboration. J Exp Bot 73, 3308-3318.
DOI URL |
| [19] |
Hase Y, Fujioka S, Yoshida S, Sun GQ, Umeda M, Tanaka A (2005). Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J Exp Bot 56, 1263-1268.
DOI URL |
| [20] |
Hase Y, Tanaka A, Baba T, Watanabe H (2000). FRL1 is required for petal and sepal development in Arabidopsis. Plant J 24, 21-32.
PMID |
| [21] |
Huang D, Li XW, Sun M, Zhang TX, Pan HT, Cheng TR, Wang J, Zhang QX (2016). Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front Plant Sci 7, 1633.
PMID |
| [22] |
Kalisz S, Ree RH, Sargent RD (2006). Linking floral symmetry genes to breeding system evolution. Trends Plant Sci 11, 568-573.
PMID |
| [23] |
Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322, 1116-1119.
DOI PMID |
| [24] |
Laitinen RAE, Broholm S, Albert VA, Teeri TH, Elomaa P (2006). Patterns of MADS-box gene expression mark flower-type development in Gerbera hybrida (Asteraceae). BMC Plant Biol 6, 11.
PMID |
| [25] |
Li H, Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760.
DOI PMID |
| [26] |
Li ZQ, Xu YH (2022). Bulk segregation analysis in the NGS era: a review of its teenage years. Plant J 109, 1355-1374.
DOI URL |
| [27] |
Liu SZ, Yeh CT, Tang HM, Nettleton D, Schnable PS (2012). Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7, e36406.
DOI URL |
| [28] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297-1303.
DOI PMID |
| [29] |
Michelmore RW, Paran I, Kesseli RV (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88, 9828-9832.
DOI PMID |
| [30] |
Ohno CK, Reddy GV, Heisler MGB, Meyerowitz EM (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111-1122.
DOI URL |
| [31] |
Panero JL, Freire SE, Ariza EL, Crozier BS, Barboza GE, Cantero JJ (2014). Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Mol Phylogen Evol 80, 43-53.
DOI URL |
| [32] |
Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E (2007). Evolution and development of inflorescence architectures. Science 316, 1452-1456.
DOI PMID |
| [33] |
Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E (2013). JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol 11, e1001550.
DOI URL |
| [34] |
Shen CZ, Chen J, Zhang CJ, Rao GY, Guo YP (2021). Dysfunction of CYC2g is responsible for the evolutionary shift from radiate to disciform flowerheads in the Chrysanthemum group (Asteraceae: Anthemideae). Plant J 106, 1024-1038.
DOI URL |
| [35] |
Song XB, Gao K, Fan GX, Zhao XG, Liu ZL, Dai SL (2018a). Quantitative classification of the morphological traits of ray florets in large-flowered chrysanthemum. HortScience 53, 1258-1265.
DOI URL |
| [36] |
Song XB, Xu YH, Gao K, Fan GX, Zhang F, Deng CY, Dai SL, Huang H, Xin HG, Li YY (2020). High-density genetic map construction and identification of loci controlling flower-type traits in chrysanthemum (Chrysanthemum × morifolium Ramat.). Hortic Res 7, 108.
DOI |
| [37] |
Song XB, Zhao XG, Fan GX, Gao K, Dai SL, Zhang MM, Ma CF, Wu XY (2018b). Genetic analysis of the corolla tube merged degree and the relative number of ray florets in chrysanthemum (Chrysanthemum × morifolium Ramat.). Sci Hortic 242, 214-224.
DOI URL |
| [38] |
Tähtiharju S, Rijpkema AS, Vetterli A, Albert VA, Teeri TH, Elomaa P (2012). Evolution and diversification of the CYC/TB1 gene family in Asteraceae—a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol Biol Evol 29, 1155-1166.
DOI PMID |
| [39] |
Wang QJ, Zhang XN, Lin SN, Yang SZ, Yan XL, Bendahmane M, Bao MZ, Fu XP (2020). Mapping a double flower phenotype-associated gene DcAP2L in Dianthus chinensis. J Exp Bot 71, 1915-1927.
DOI URL |
| [40] |
Wyatt R (1982). Inflorescence architecture: how flower number, arrangement, and phenology affect pollination and fruit-set. Am J Bot 69, 585-594.
DOI URL |
| [41] |
Zhao YF, Zhang T, Broholm SK, Tähtiharju S, Mouhu K, Albert VA, Teeri TH, Elomaa P (2016). Evolutionary co-option of floral meristem identity genes for patterning of the flower-like Asteraceae inflorescence. Plant Physiol 172, 284-296.
DOI PMID |
| [42] |
Zheng GH, Wei W, Li YP, Kan LJ, Wang FX, Zhang X, Li F, Liu ZC, Kang CY (2019). Conserved and novel roles of miR164-CUC2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytol 224, 480-492.
DOI URL |
| [1] | Zhigang Yang, Pengcheng Zhang, Haiwen Chang, Liru Kang, Yi Zuo, Haoxin Xiang, Fengying Han. Genetic Diversity Analysis of Pepper Germplasms Based on Morphological Traits and SSR Markers [J]. Chinese Bulletin of Botany, 2025, 60(2): 218-234. |
| [2] | Yi Zuo, Hongbing Liu, Zhigang Yang, Bin Li, Haoxin Xiang, Chunzhen Zhu, Lei Wang. Identification of Sex Determination Molecular Marker Based on Genome-wide Association Study of Idesia polycarpa [J]. Chinese Bulletin of Botany, 2024, 59(3): 414-421. |
| [3] | Hongwei Li, Qi Zheng, Bin Li, Zhensheng Li. Research Progress on the Aspects of Molecular Breeding of Tall Wheatgrass [J]. Chinese Bulletin of Botany, 2022, 57(6): 792-801. |
| [4] | Wenqing Tan, Jun Chen, Hongwei Cai. Recent Progress in Biology of Genus Lolium [J]. Chinese Bulletin of Botany, 2022, 57(6): 802-813. |
| [5] | Xiangxiang Chen, Zhongshuai Gai, Juntuan Zhai, Jindong Xu, Peipei Jiao, Zhihua Wu, Zhijun Li. Genetic diversity and construction of core conservation units of the natural populations of Populus euphratica in Northwest China [J]. Biodiv Sci, 2021, 29(12): 1638-1649. |
| [6] | Yaqin Wang, Ludan Wei, Wenjing Wang, Baojun Liu, Chunling Zhang, Junwei Zhang, Yanhong He. The Establishment and Optimization of a Regeneration System for Marigold (Tagetes erecta) [J]. Chinese Bulletin of Botany, 2020, 55(6): 749-759. |
| [7] | Yuanyuan Li, Chaonan Liu, Rong Wang, Shuixing Luo, Shouqian Nong, Jingwen Wang, Xiaoyong Chen. Applications of molecular markers in conserving endangered species [J]. Biodiv Sci, 2020, 28(3): 367-375. |
| [8] | Yahong Zhang, Huixia Jia, Zhibin Wang, Pei Sun, Demei Cao, Jianjun Hu. Genetic diversity and population structure of Populus yunnanensis [J]. Biodiv Sci, 2019, 27(4): 355-365. |
| [9] | Jie Yang, Jia He, Danbi Wang, En Shi, Wenyu Yang, Qifang Geng, Zhongsheng Wang*. Progress in research and application of InDel markers [J]. Biodiv Sci, 2016, 24(2): 237-243. |
| [10] | Qi Guo, Dalong Guo, Lili Guo, Lin Zhang, Xiaogai Hou. Application of Simple Sequence Repeat Molecular Markers in the Study of Tree Peony [J]. Chinese Bulletin of Botany, 2015, 50(5): 652-664. |
| [11] | Dangni Zhang, Lianming Zheng, Jinru He, Wenjing Zhang, Yuanshao Lin, Yang Li. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification [J]. Biodiv Sci, 2015, 23(1): 50-60. |
| [12] | Xi Zhang, Xiaogai Hou, Dalong Guo, Chengwei Song, Yabin Duan. iPBS-PCR Used for Cloning and Analysis of Long Terminal Repeat Transposons in Tree Peony (Paeonia) [J]. Chinese Bulletin of Botany, 2014, 49(3): 322-330. |
| [13] | Wei Zhou,Hong Wang. Pollen dispersal analysis using DNA markers [J]. Biodiv Sci, 2014, 22(1): 97-108. |
| [14] | Wen Liu, Hong Liang, Biren Yang, Jiewen Lin, Yanji Hu. Short RNAs, potential novel molecular markers for higher plants [J]. Biodiv Sci, 2012, 20(1): 86-93. |
| [15] | Zhaoxia Cui, Huan Zhang, Linsheng Song, Feng You. Genetic diversity of marine animals in China: a summary and prospec- tiveness [J]. Biodiv Sci, 2011, 19(6): 815-833. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||