

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (6): 782-792.DOI: 10.11983/CBB17258 cstr: 32102.14.CBB17258
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Luo Junjie1,2, Wang Ying1,2, Shang Hui1,3, Zhou Xile4, Wei Hongjin1,3, Huang Sunan2, Gu Yufeng1,2, Jin Dongmei1,3, Dai Xiling2, Yan Yuehong1,3,*( )
)
Received:2017-12-29
Online:2018-11-01
Published:2018-12-05
Contact:
Yan Yuehong
Luo Junjie, Wang Ying, Shang Hui, Zhou Xile, Wei Hongjin, Huang Sunan, Gu Yufeng, Jin Dongmei, Dai Xiling, Yan Yuehong. Phylogeny and Systematics of the Genus Microlepia (Dennstaedtiaceae) based on Palynology and Molecular Evidence[J]. Chinese Bulletin of Botany, 2018, 53(6): 782-792.
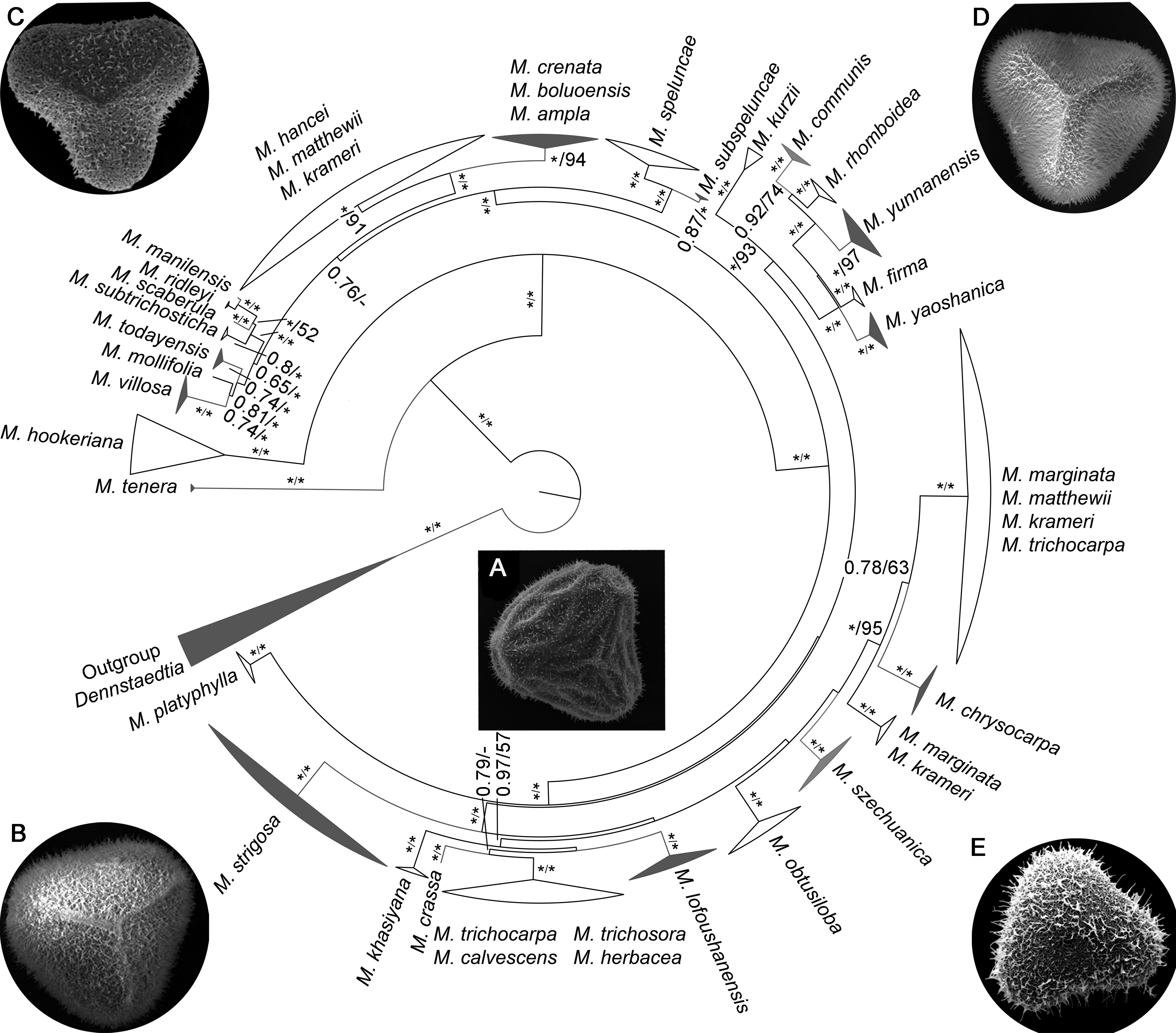
Figure 1 Bayesian inference phylogeny of Microlepia derived from the combined data (rbcL, trnL-F, psbA-trnH and rps4) Values beside each branch represent bootstrap support for parsimony, Bayesian posterior probabilities and maximum likelihood (≥50%); * means 1 or 100; - means <0.5 or 50%.
| [1] | 曹建国, 于晶, 王全喜 (2007). 中国蕨类植物孢子的形态VII. 桫椤科. 云南植物研究 29, 7-12. |
| [2] | 陈焕镛 (1964). 海南植物志, 第1卷. 北京: 科学出版社. pp. 46-52. |
| [3] | 广西植物研究所 (2013). 广西植物志. 南宁: 广西科学技术出版社. pp. 96-108. |
| [4] | 孔宪需 (1988). 四川植物志, 第6卷. 蕨类植物门. 成都: 四川科学技术出版社. pp. 183-190. |
| [5] |
刘红梅, 张宪春, 曾辉 (2009). DNA序列在蕨类分子系统学研究中的应用. 植物学报 44, 143-158.
DOI URL |
| [6] | 牟善杰 (2010). 碗蕨科鳞盖蕨属专论研究. 博士论文. 台北: 台湾师范大学. pp. 1-198. |
| [7] | 秦仁昌 (1959). 中国植物志, 第2卷. 北京: 科学出版社. pp. 207-246. |
| [8] | 王培善, 王筱英 (2001). 贵州蕨类植物志. 贵阳: 贵州科技出版社. pp. 433-442. |
| [9] | 王全喜, 陈立群, 包文美 (1997). 中国金毛裸蕨属植物孢子形态的研究. 西北植物学报 17(5), 44-47. |
| [10] | 王全喜, 戴锡玲 (2010). 中国水龙骨目(真蕨目)植物孢子形态的研究. 北京: 科学出版社. pp. 1-25. |
| [11] |
王全喜, 于晶 (2003). 扫描电镜下真蕨目孢子表面纹饰的分类. 云南植物研究 25, 313-320.
DOI URL |
| [12] | 吴德邻 (2006). 广东植物志, 第7卷. 广州: 广东科技出版社. pp. 81-85. |
| [13] | 吴征镒 (2006). 云南植物志, 第12卷. 北京: 科学出版社. pp. 216-230. |
| [14] | 杨鲁红 (2012). 中国石韦属植物系统分类学研究. 硕士论文. 昆明: 云南大学. pp. 1-142. |
| [15] |
张宪春, 卫然, 刘红梅, 何丽娟, 王丽, 张钢民 (2013). 中国现代石松类和蕨类的系统发育与分类系统. 植物学报 48, 119-137.
DOI URL |
| [16] | 张玉龙, 席以珍, 张金谈, 高桂珍, 杜乃秋, 孙湘君, 孔昭宸 (1976). 中国蕨类植物孢子形态. 北京: 科学出版社. pp. 1-112. |
| [17] |
Akaike H (1974). A new look at the statistical model identification.IEEE Trans Automat Contr 19, 716-723.
DOI URL |
| [18] | Burland TG (2000). Dnastar’s lasergene sequence analysis software. In: Misener S, Krawetz SA, eds. Bioinformatics Methods and Protocols. Methods in Molecular BiologyTM, Vol. 132. Totowa, NJ: Humana Press. pp. 71-91. |
| [19] |
Denk T, Grimm GW (2009). Significance of pollen characteristics for infrageneric classification and phylogeny in Quercus(Fagaceae). Int J Plant Sci 170, 926-940.
DOI URL |
| [20] |
Ebihara A, Nitta JH, Ito M (2010). Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan.PLoS One 5, e15136.
DOI URL PMID |
| [21] | Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT.Nucl Acids Sym Ser 41, 95-98. |
| [22] | Hasebe M, Omori T, Nakazawa M, Sano T, Kato M, Iwatsuki K (1994). rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proc Natl Acad Sci USA 91, 5730-5734. |
| [23] | Huang TC (1981). Spore Flora of Taiwan (Pteridophyta). Taiwan: National Taiwan University. pp. 52-53. |
| [24] | Kramer KU (1990). Dennstaedtiaceae. In: Kramer KU, Green PS, eds. The Families and Genera of Vascular Plants, Vol. I. Pteridophytes and Gymnosperms. Berlin: Springer-Verlag. pp. 81-94. |
| [25] |
Lehtonen S, Wahlberg N, Christenhusz MJM (2012). Diversification of lindsaeoid ferns and phylogenetic uncertainty of early polypod relationships.Bot J Linn Soc 170, 489-503.
DOI URL |
| [26] |
Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, Windham MD, Pryer KM (2011). rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS One 6, e26597.
DOI URL PMID |
| [27] | Miller MA, Pfeiffer W, Schwartz T (2010). Creating the cipres science gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE). New Orleans: IEEE. pp. 1-8. |
| [28] | Moran RC, Hanks JG, Rouhan G (2007). Spore morpho- logy in relation to phylogeny in the fern genus Elaphoglossum(Dryopteridaceae). Int J Plant Sci 168, 905-929. |
| [29] | Nakato N, Ebihara A (2011). Chromosome number of Microlepia hookeriana (Dennstaedtiaceae) and chromosome number evolution in the genus Microlepia. Bull Natl Mus Nat Sci Ser B 37, 75-78. |
| [30] |
Posada D (2008). Jmodeltest: phylogenetic model averaging.Mol Biol Evol 25, 1253-1256.
DOI URL |
| [31] |
Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R (2004). Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences.Am J Bot 91, 1582-1598.
DOI URL PMID |
| [32] |
Pryer KM, Smith AR, Hunt JS, Dubuisson JY (2001). rbcL data reveal two monophyletic groups of filmy ferns(Filicopsida: Hymenophyllaceae). Am J Bot 88, 1118-1130.
DOI URL PMID |
| [33] | Rambaut A, Drummond AJ (2007). Tracer v1.4. . |
| [34] |
Ronquist F, Huelsenbeck JP (2003). Mrbayes 3: Bayesian phylogenetic inference under mixed models.Bioinforma- tics 19, 1572-1574.
DOI URL |
| [35] |
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Syst Biol 61, 539-542.
DOI URL PMID |
| [36] | Schuettpelz E, Korall P, Pryer KM (2006). Plastid atpA data provide improved support for deep relationships among ferns. Taxon 55, 897-906. |
| [37] |
Schuettpelz E, Pryer KM (2007). Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes.Taxon 56, 1037-1050.
DOI URL |
| [38] |
Schwarz G (1978). Estimating the dimension of a model.Ann Statist 6, 461-464.
DOI URL |
| [39] |
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu WS, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005). The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis.Am J Bot 92, 142-166.
DOI URL |
| [40] |
Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG (2006). A classification for extant ferns.Taxon 55, 705-731.
DOI URL |
| [41] | Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B (1997). Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Syst Evol 204, 109-123. |
| [42] | Swofford DL (2003). Paup*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. |
| [43] |
Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA.Plant Mol Biol 17, 1105-1109.
DOI URL |
| [44] | Tagawa M (1952). Fern miscellany (6).J Jpn Bot 27, 213-218. |
| [45] |
Tate JA, Simpson BB (2003). Paraphyly of Tarasa(Malvaceae) and diverse origins of the polyploid species. Syst Bot 28, 723-737.
DOI URL |
| [46] |
The Pteridophyte Phylogeny Group (2016). A community-derived classification for extant lycophytes and ferns.J Syst Evol 54, 563-603.
DOI URL |
| [47] | Tryon AF, Lugardon B (1991). Spores of the Pteridophyta. Berlin: Springer-Verlag. pp. 1-279. |
| [48] |
Wang FG, Liu HM, He CM, Yang DM, Xing FW (2015). Taxonomic and evolutionary implications of spore ornamentation in Davalliaceae.J Syst Evol 53, 72-81.
DOI URL |
| [49] |
Wolf PG (1995). Phylogenetic analyses of rbcL and nuclear ribosomal RNA gene sequences in Dennstaedtiaceae. Am Fern J 85, 306-327.
DOI URL |
| [50] |
Wolf PG (1997). Evaluation of atpB nucleotide sequences for phylogenetic studies of ferns and other pteridophytes. Am J Bot 84, 1429-1440.
DOI URL PMID |
| [51] |
Xu Y, Hu CM, Hao G (2016). Pollen morphology of Androsace(Primulaceae) and its systematic implications. J Syst Evol 54, 48-64.
DOI URL |
| [52] | Yan YH, Qi XP, Zhang XC (2013). Dennstaedtiaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China, Vol. 2-3. Pteridophytes. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. pp. 147-168. |
| [53] | Yanez A, Marquez GJ, Morbelli MA (2016). Palynological analysis of Dennstaedtiaceae taxa from the paranaense phytogeographic province that produce trilete spores II: Microlepia speluncae and Pteridium arachnoideum. An Acad Bras Ciênc 88, 877-890. |
| [54] |
Yuan Y, Fu L, Ma CY (2012). Microlepia boluoensis sp. nov.(Dennsteadtiaceae) from Guangdong, China. Nord J Bot 30, 168-173.
DOI URL |
| [55] |
Zhou XM, Zhang LB (2015). A classification of Selaginella(Selaginellaceae) based on molecular (chloroplast and nuclear), macromorphological, and spore features. Taxon 64, 1117-1140.
DOI URL |
| [1] | Xuan Zhou, Shengfang Zhang, Ning Liu, Yujie Lu, Sizhu Zheng, Xiaojun Yang, Yuanyuan Lu, Meike Liu, Ming Bai. Revision of the systematic status and update of Latin-English-Chinese catalogue of storage beetles [J]. Biodiv Sci, 2025, 33(4): 24238-. |
| [2] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| [3] | Ke Wang, Mingjun Zhao, Lei Cai. Annual review on fungi nomenclature novelties in China and around the world (2023) [J]. Biodiv Sci, 2024, 32(11): 24361-. |
| [4] | Xinyu Li, Yaxuan Zhang, Meichen Yan, Ruihan Yang, Xiaoqing Zhang, Zhiyuan Yao. New taxa of extant spiders (Araneae) from the world in 2023 [J]. Biodiv Sci, 2024, 32(11): 24181-. |
| [5] | Dekui He, Jinnan Chen, Liuyong Ding, Yiyang Xu, Junhao Huang, Xiaoyun Sui. The status and distribution pattern of fish diversity in the Yarlung Tsangpo River [J]. Biodiv Sci, 2024, 32(11): 24143-. |
| [6] | Hong Du. “Species” versus “individuals”: Which is the right target for biodiversity conservation? [J]. Biodiv Sci, 2023, 31(8): 23140-. |
| [7] | Wang Lulu, Yang Zhi, Yang Yong. Plant Ultra-barcoding Using Herbariomics [J]. Chinese Bulletin of Botany, 2023, 58(5): 831-842. |
| [8] | Huiyin Song, Zhengyu Hu, Guoxiang Liu. Assessing advances in taxonomic research on Chlorellaceae (Chlorophyta) [J]. Biodiv Sci, 2023, 31(2): 22083-. |
| [9] | Xuan Zhou, Yiping Luo, Zi Jin, Yuqing Qiao, Yiyao Zhang, Lulu Li, Yuanyuan Lu, Ning Liu, Meike Liu, Ming Bai. The world new taxa for extant Coleoptera in 2022 [J]. Biodiv Sci, 2023, 31(10): 23202-. |
| [10] | Ke Wang, Lei Cai. Annual review on nomenclature novelties of fungi in the world (2022) [J]. Biodiv Sci, 2023, 31(10): 23176-. |
| [11] | Ruihan Yang, Meichen Yan, Ludan Zhang, Hongxin Liu, Joseph KH Koh, Qiaoqiao He, Zhiyuan Yao. New taxa of spiders (Araneae) from the world in 2022 [J]. Biodiv Sci, 2023, 31(10): 23175-. |
| [12] | Ke Liu, Sicheng Han, He Yu, Shu-Jin Luo. The evolutionary genetics, taxonomy, and conservation of the Chinese mountain cat [J]. Biodiv Sci, 2022, 30(9): 22396-. |
| [13] | Ludan Zhang, Ying Lu, Chang Chu, Qiaoqiao He, Zhiyuan Yao. New spider taxa of the world in 2021 [J]. Biodiv Sci, 2022, 30(8): 22163-. |
| [14] | Ke Wang, Lei Cai. Annual review on nomenclature novelties of fungi in the world (2021) [J]. Biodiv Sci, 2022, 30(8): 22277-. |
| [15] | Chen Lin, Bing Zhang, Liang Wang, Ding Yang. New taxa of Diptera from China in 2021 [J]. Biodiv Sci, 2022, 30(8): 22192-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||