

点击化学反应在植物细胞标记中的应用
收稿日期: 2022-10-25
录用日期: 2023-02-28
网络出版日期: 2023-03-24
基金资助
国家自然科学基金(91954202);国家自然科学基金(31871349);北京林业大学科技创新计划(2019JQ03003)
The Application of Click Chemistry Reactions in Plant Cell Labeling
Received date: 2022-10-25
Accepted date: 2023-02-28
Online published: 2023-03-24
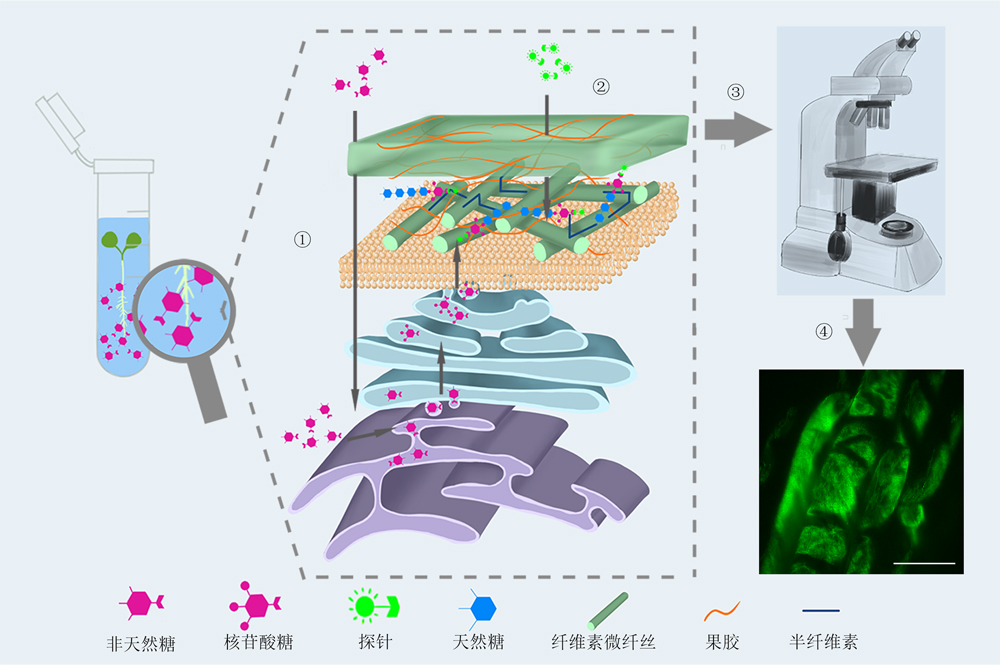
点击化学又称“链接化学”或“速配结合式组合化学”。其可通过碳-杂原子键(C-X-C)连接产生出诸多功能强大、高度可靠且具较强特异性的反应, 是一种快速合成大量化合物的新方法。近几年, 点击化学在药物开发、新材料合成、材料表面功能化修饰和生物大分子标记等方面取得了较大进展。2022年, 点击化学的开拓者获得了诺贝尔化学奖。该文简要介绍点击化学的原理和反应类型, 重点总结其在标记生物大分子上的研究进展, 特别是在植物细胞壁聚糖标记方面的应用, 以期为解析植物细胞壁结构、合成和动态转运机制提供新思路。
关键词: 点击化学反应; 叠氮-炔烃环加成反应; 细胞壁; 聚糖
张御格 , 袁笑妍 , 张贵芳 , 李雨健 , 殷金环 , 林金星 , 李晓娟 . 点击化学反应在植物细胞标记中的应用[J]. 植物学报, 2023 , 58(6) : 956 -965 . DOI: 10.11983/CBB22252
Click chemistry, also known as "link chemistry" or "speed-matching combinatorial chemistry", is the development of a powerful, highly reliable, and highly specific set of reactions for the rapid synthesis of new compounds through the connection of carbon-heteroatomic bonds (C-X-C). Click chemistry has been progressing greatly in drug development, new material synthesis, material surface functionalization modification and biological macromolecular labeling. Pioneers of click chemistry was awarded the Nobel Prize in Chemistry in 2022. This review briefly introduces the principles, reaction types and applications of click chemistry, summarizes the research progress of click chemistry in labeling biological macromolecules, especially in its application to plant cell wall polysaccharide labeling, and provides new ideas for the study of plant cell wall synthesis, structure and dynamic transport.

Key words: click chemistry reaction; azide-alkyne cycloaddition; cell wall; glycan
| [1] | 成波, 陈兴 (2020). 唾液酸化聚糖的化学标记和解析. 科学通报 65, 2984-2997. |
| [2] | 刘佩佩, 张耿, 李晓娟 (2021). 植物果胶的生物合成与功能. 植物学报 56, 191-200. |
| [3] | 刘玥, 尹悦佳, 梁重阳, 黄殿帅, 王阳, 刘艳芝, 窦瑶, 冯树丹, 郝东云 (2015). 3D-SIM结构照明超分辨率显微镜实现蛋白质在植物亚细胞器内的定位. 植物学报 50, 495-503. |
| [4] | 肖银燕, 袁伟娜, 刘静, 孟建, 盛奇明, 谭烨欢, 徐春香 (2020). 木葡聚糖及其在植物抗逆过程中的功能研究进展. 植物学报 55, 777-787. |
| [5] | 杨麦云, 陈鹏 (2015). 生物正交标记反应研究进展. 化学学报 73, 783-792. |
| [6] | 占方玲, 高思宇, 谢元栋, 张金铭, 李毅, 刘宁 (2020). 点击化学反应在蛋白质组学分析中的研究进展. 分析化学 48, 431-438. |
| [7] | Finn MG, Kolb HC, Fokin VV, Sharpless KB (张欣豪, 吴云东译)(2008). 点击化学——释义与目标. 化学进展 20, 1-4. |
| [8] | Agard NJ, Prescher JA, Bertozzi CR (2004). A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc 126, 15046-15047. |
| [9] | Ancajas CF, Ricks TJ, Best MD (2020). Metabolic labeling of glycerophospholipids via clickable analogs derivatized at the lipid headgroup. Chem Phys Lipids 232, 104971. |
| [10] | Anderson CT, Wallace IS, Somerville CR (2012). metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc Natl Acad Sci USA 109, 1329-1334. |
| [11] | Beatty KE, Xie F, Wang Q, Tirrell DA (2005). Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc 127, 14150-14151. |
| [12] | Beller NC, Hummon AB (2022). Advances in stable isotope labeling: dynamic labeling for spatial and temporal proteo-mic analysis. Mol Omics 18, 579-590. |
| [13] | Bidhendi AJ, Chebli Y, Geitmann A (2020). Fluorescence visualization of cellulose and pectin in the primary plant cell wall. J Microsc 278, 164-181. |
| [14] | Bird RE, Lemmel SA, Yu X, Zhou QA (2021). Bioorthogonal chemistry and its applications. Bioconjug Chem 32, 2457-2479. |
| [15] | Breugst M, Reissig HU (2020). The Huisgen reaction: milestones of the 1,3-dipolar cycloaddition. Angew Chem Int Ed Engl 59, 12293-12307. |
| [16] | Chang XL, Chen LY, Liu BN, Yang ST, Wang HF, Cao AN, Chen CY (2022). Stable isotope labeling of nanomaterials for biosafety evaluation and drug development. Chin Chem Lett 33, 3303-3314. |
| [17] | Cheng B, Tang Q, Zhang C, Chen X (2021). Glycan labeling and analysis in cells and in vivo. Annu Rev Anal Chem (Palo Alto Calif) 14, 363-387. |
| [18] | Depmeier H, Hoffmann E, Bornewasser L, Kath-Schorr S (2021). Strategies for covalent labeling of long RNAs. Chembiochem 22, 2826-2847. |
| [19] | Devaraj NK, Finn MG (2021). Introduction: click chemistry. Chem Rev 121, 6697-6698. |
| [20] | Devree BT, Steiner LM, G?azowska S, Ruhnow F, Her-burger K, Persson S, Mravec J (2021). Current and future advances in fluorescence-based visualization of plant cell wall components and cell wall biosynthetic machineries. Biotechnol Biofuels 14, 78. |
| [21] | Dumont M, Lehner A, Vauzeilles B, Malassis J, Marchant A, Smyth K, Linclau B, Baron A, Mas Pons J, Ander-son CT, Schapman D, Galas L, Mollet JC, Lerouge P (2016). Plant cell wall imaging by metabolic click-mediated labeling of rhamnogalacturonan II using azido 3-deoxy-D-manno-oct-2-ulosonic acid. Plant J 85, 437-447. |
| [22] | Fantoni NZ, El-Sagheer AH, Brown T (2021). A hitchhiker’s guide to click-chemistry with nucleic acids. Chem Rev 121, 7122-7154. |
| [23] | Gothelf KV, J?rgensen KA (1998). Asymmetric 1,3-dipolar cycloaddition reactions. Chem Rev 98, 863-910. |
| [24] | Hoogenboom J, Berghuis N, Cramer D, Geurts R, Zuilhof H, Wennekes T (2016). Direct imaging of glycans in Arabidopsis roots via click labeling of metabolically incorporated azido-monosaccharides. BMC Plant Biol 16, 220. |
| [25] | Huisgen R (1963). 1,3-dipolar cycloadditions. Past and future. Angew Chem Int Ed Engl 2, 565-598. |
| [26] | Jao CY, Roth M, Welti R, Salic A (2009). Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc Natl Acad Sci USA 106, 15332-15337. |
| [27] | Jao CY, Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA 105, 15779-15784. |
| [28] | John CJ, Carolyn RB (2010). Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev 39, 1272-1279. |
| [29] | Kim E, Koo H (2019). Biomedical applications of copper- free click chemistry: in vitro, in vivo, and ex vivo. Chem Sci, 10, 7835-7851. |
| [30] | Kolb HC, Finn MG, Sharpless KB (2001). Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl 40, 2004-2021. |
| [31] | Kuerschner L, Thiele C (2022). Tracing lipid metabolism by alkyne lipids and mass spectrometry: the state of the art. Front Mol Biosci 9, 880559. |
| [32] | Li L, Zhang ZY (2016). Development and applications of the copper-catalyzed azide-alkyne cycloaddition (CuAAC) as a bioorthogonal reaction. Molecules 21, 1393. |
| [33] | Lion C, Simon C, Huss B, Blervacq AS, Tirot L, Toybou D, Spriet C, Slomianny C, Guerardel Y, Hawkins S, Biot C (2017). BLISS: a bioorthogonal dual-labeling strategy to unravel lignification dynamics in plants. Cell Chem Biol 24, 326-338. |
| [34] | Ming X, Leonard P, Heindl D, Seela F (2008). Azide-alkyne "click" reaction performed on oligonucleotides with the universal nucleoside 7-octadiynyl-7-deaza-2'-deoxyinosine. Nucleic Acids Symp Ser 52, 471-472. |
| [35] | Neef AB, Luedtke NW (2014). An azide-modified nucleoside for metabolic labeling of DNA. Chembiochem 15, 789-793. |
| [36] | Neef AB, Pernot L, Schreier VN, Scapozza L, Luedtke NW (2015). A bioorthogonal chemical reporter of viral infection. Angew Chem Int Ed Engl 54, 7911-7914. |
| [37] | Paper JM, Mukherjee T, Schrick K (2018). Bioorthogonal click chemistry for fluorescence imaging of choline phospholipids in plants. Plant Methods 14, 31. |
| [38] | Parker CG, Pratt MR (2020). Click chemistry in proteomic investigations. Cell 180, 605-632. |
| [39] | Prescher JA, Bertozzi CR (2005). Chemistry in living sys-tems. Nat Chem Biol 1, 13-21. |
| [40] | Rodríguez DF, Moglie Y, Ramírez-Sarmiento CA, Singh SK, Dua K, Zacconi FC (2022). Bio-click chemistry: a bridge between biocatalysis and click chemistry. RSC Adv 12, 1932-1949. |
| [41] | Ropitaux M, Hays Q, Baron A, Fourmois L, Boulogne I, Vauzeilles B, Lerouge P, Mollet JC, Lehner A (2022). Dynamic imaging of cell wall polysaccharides by metabolic click-mediated labeling of pectins in living elongating cells. Plant J 110, 916-924. |
| [42] | Rostovtsev VV, Green LG, Fokin VV, Sharpless KB (2002). A stepwise Huisgen cycloaddition process: copper (I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl 41, 2596-2599. |
| [43] | Takayama Y, Kusamori K, Nishikawa M (2019). Click chemistry as a tool for cell engineering and drug delivery. Molecules 24, 172. |
| [44] | Teramoto H, Kojima K (2015). Incorporation of methionine analogues into Bombyx mori silk fibroin for click modifica-tions. Macromol Biosci 15, 719-727. |
| [45] | Tobimatsu Y (2017). A “double click” for illuminating plant cell walls. Cell Chem Biol 24, 246-247. |
| [46] | Truong L, Ferré-D'Amaré AR (2019). From fluorescent proteins to fluorogenic RNAs: tools for imaging cellular macromolecules. Protein Sci 28, 1374-1386. |
| [47] | Turner MA, Lwin TM, Amirfakhri S, Nishino H, Hoffman RM, Yazaki PJ, Bouvet M (2021). The use of fluorescent anti-CEA antibodies to label, resect and treat cancers: a review. Biomolecules 11, 1819. |
| [48] | Wang JG, Zhang JB, Lee YM, Ng S, Shi Y, Hua ZC, Lin QS, Shen HM (2017). Nonradioactive quantification of autophagic protein degradation with L-azidohomoalanine labeling. Nat Protoc 12, 279-288. |
| [49] | Wittig G, Krebs A (1961). Zur existenz niedergliedriger cycloalkine, 1. Chem Ber 94, 3260-3275. |
| [50] | Wu J, Yu YH (2015). Recent progress on application of “click” chemistry in labeling of biomolecules. J Jianghan Univ (Nat Sci Ed) 43, 138-145. |
| [51] | Xiong HT, Zou HY, Liu H, Wang M, Duan LL (2021). Sur-face functionalization of a γ-graphyne-like carbon material via click chemistry. Chem Asian J 16, 922-925. |
| [52] | Yao TT, Xu XW, Huang R (2021). Recent advances about the applications of click reaction in chemical proteomics. Molecules 26, 5368. |
| [53] | Yoon HY, Lee D, Lim DK, Koo H, Kim K (2022). Cop-per-free click chemistry: applications in drug delivery, cell tracking, and tissue engineering. Adv Mater 34, 2107192. |
| [54] | Zhu YT, Chen X (2017). Expanding the scope of metabolic glycan labeling in Arabidopsis thaliana. Chembiochem 18, 1286-1296. |
| [55] | Zhu YT, Wu J, Chen X (2016). Metabolic labeling and imaging of N-linked glycans in Arabidopsis thaliana. Angew Chem Int Ed Engl 55, 9301-9305. |
| [56] | Zou J, Dong XY, Li YL, Tong SQ, Wang JW, Liao MX, Huang GF (2019). Deep sequencing identification of differentially expressed miRNAs in the spinal cord of resinifera-toxin-treated rats in response to electroacupuncture. Neurotox Res 36, 387-395. |
/
| 〈 |
|
〉 |