

收稿日期: 2019-10-24
录用日期: 2019-12-31
网络出版日期: 2020-01-03
基金资助
国家自然科学基金(No.31670248);国家自然科学基金(No.31970276)
Protocols for Analyzing Plant Phospho-proteins
Received date: 2019-10-24
Accepted date: 2019-12-31
Online published: 2020-01-03
朱丹,曹汉威,李媛,任东涛 . 植物蛋白磷酸化的检测方法[J]. 植物学报, 2020 , 55(1) : 76 -82 . DOI: 10.11983/CBB19208
Protein phosphorylation is one of the important protein posttranslational modifications that is involved in the regulation of most cellular processes in plants. Protein kinases catalyze the phosphorylation by transferring the phosphate group in ATP to the substrate proteins. The phosphate is usually covalently linked to the hydroxyl group of specific amino acid residues in the substrates by an ester bond. The mostly studied phosphorylation sites are serine, threonine, and tyrosine residues. Here, we present protocols and related tips for the in vitro and in vivo protein phosphorylation assays.
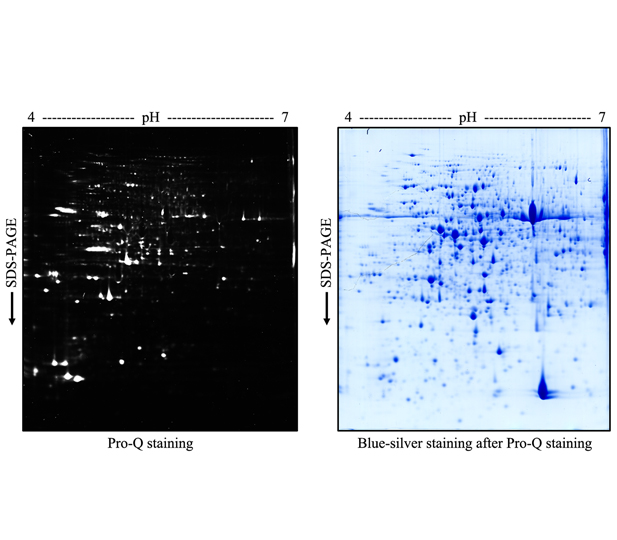
Key words: protein phosphorylation; protein kinase; plant
| [1] | Agrawal GK, Thelen JJ (2005). Development of a simplified, economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics 5, 4684-4688. |
| [2] | Agrawal GK, Thelen JJ (2006). Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics 5, 2044-2059. |
| [3] | Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633-1649. |
| [4] | Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG (2004). Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327-1333. |
| [5] | Chao Q, Liu XY, Mei YC, Gao ZF, Chen YB, Qian CR, Hao YB, Wang BC (2014). Light-regulated phosphorylation of maize phosphoenolpyruvate carboxykinase plays a vital role in its activity. Plant Mol Biol 85, 95-105. |
| [6] | Chen MJ, Dixon JE, Manning G (2017). Genomics and evolution of protein phosphatases. Sci Signal 10, eaag1796. |
| [7] | de la Fuente van Bentem S, Hirt H (2007). Using phosphoproteomics to reveal signaling dynamics in plants. Trends Plant Sci 12, 404-411. |
| [8] | Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM (2002). Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 20, 301-305. |
| [9] | Fischer EH, Krebs EG (1955). Conversion of phosphorylase B to phosphorylase A in muscle extracts. J Biol Chem 216, 121-132. |
| [10] | Frost DC, Li LJ (2014). Recent advances in mass spectrometry-based glycoproteomics. Adv Protein Chem Struct Biol 95, 71-123. |
| [11] | Hubbard MJ, Cohen P (1993). On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci 18, 172-177. |
| [12] | Ke YQ, Han GQ, He HQ, Li JX (2009). Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem Biophys Res Commun 379, 133-138. |
| [13] | Khan M, Takasaki H, Komatsu S (2005). Comprehensive phosphoproteome analysis in rice and identification of phosphoproteins responsive to different hormones/stresses. J Proteome Res 4, 1592-1599. |
| [14] | Kim HS, Fernandes G, Lee CW (2016). Protein phosphatases involved in regulating mitosis: facts and hypotheses. Mol Cell 39, 654-662. |
| [15] | Kinoshita E, Kinoshita-Kikuta E, Koike T (2007). Specific recognition and detection of phosphorylated proteins using characteristics of metal ion. Yakugaku Zasshi 127, 1897-1913. |
| [16] | Kosako H, Nagano K (2011). Quantitative phosphoproteomics strategies for understanding protein kinase-mediated signal transduction pathways. Expert Rev Proteomics 8, 81-94. |
| [17] | Krupa A, Preethi G, Srinivasan N (2004). Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J Mol Biol 339, 1025-1039. |
| [18] | Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. |
| [19] | Laugesen S, Bergoin A, Rossignol M (2004). Deciphering the plant phosphoproteome: tools and strategies for a challenging task. Plant Physiol Biochem 42, 929-936. |
| [20] | Peck SC (2003). Early phosphorylation events in biotic stress. Curr Opin Plant Biol 6, 334-338. |
| [21] | Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008). Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7, 1019-1030. |
| [22] | Wang PC, Zhao Y, Li ZP, Hsu CC, Liu X, Fu LW, Hou YJ, Du YY, Xie SJ, Zhang CG, Gao JH, Cao MJ, Huang XS, Zhu YF, Tang K, Wang XG, Tao WA, Xiong Y, Zhu JK (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69, 100-112. |
| [23] | Whiteman SA, Nühse TS, Ashford DA, Sanders D, Maathuis FJ (2008). A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J 56, 146-156. |
| [24] | Wu CF, Wang RN, Liang QJ, Liang JJ, Li WK, Jung SY, Qin J, Lin SH, Kuang J (2010). Dissecting the M phase- specific phosphorylation of serine-proline or threonine- proline motifs. Mol Biol Cell 21, 1470-1481. |
| [25] | Yang C, Wang ZG, Zhu PF (2004). Recent advances of protein phosphorylation in proteome. Prog Physiol Sci 35, 119-124. |
| [26] | Yin XJ, Wang X, Komatsu S (2018). Phosphoproteomics: protein phosphorylation in regulation of seed germination and plant growth. Curr Protein Pept Sci 19, 401-412. |
| [27] | Zhang X, Cui YN, Yu M, Su BD, Gong W, Balu?ka F, Komis G, Samaj J, Shan XY, Lin JX (2019). Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol 181, 480-498. |
| [28] | Zhu WG (2017). Regulation of p53 acetylation. Sci China Life Sci 60, 321-323. |
/
| 〈 |
|
〉 |