

药用野生稻GBSS基因的系统发育及组织特异性表达
收稿日期: 2018-04-26
录用日期: 2018-12-10
网络出版日期: 2018-12-10
基金资助
国家自然科学基金(31570218)
Phylogeny and Tissue-specific Expression of the GBSS Genes in Oryza officinalis
Received date: 2018-04-26
Accepted date: 2018-12-10
Online published: 2018-12-10
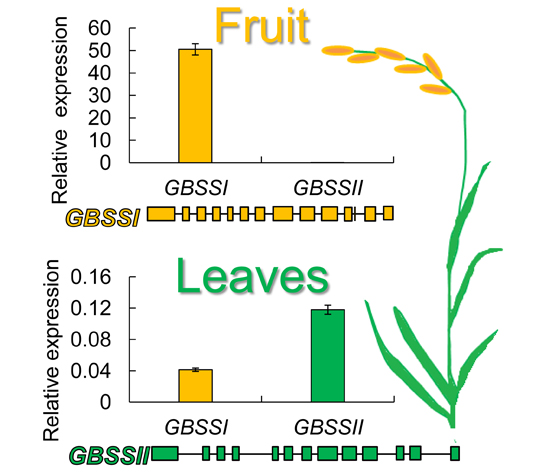
淀粉作为主要的碳水化合物在储藏能量方面发挥至关重要的作用。颗粒结合型淀粉合酶(GBSS)与直链淀粉的合成息息相关。尽管该酶的编码基因已在许多栽培植物中被分离和确定, 但有关它们在作物野生近缘种中的序列分歧和表达的研究却相对较少。该研究以药用野生稻(Oryza officinalis)为研究对象, 定性和定量地分析了GBSS编码基因的序列特点、与其它植物同源基因的进化关系以及在叶和种子中的表达情况。系统发育分析表明, 该酶在禾本科植物中分别由GBSSI和GBSSII基因编码。在药用野生稻中, 这2种基因所编码蛋白的氨基酸序列一致性为62%, 并且它们在不同器官内呈现时空分化表达, 其中GBSSI在种子中超强表达, GBSSII则主要在叶片表达。
张霞 , 景翔 , 周光才 , 包颖 . 药用野生稻GBSS基因的系统发育及组织特异性表达[J]. 植物学报, 2019 , 54(3) : 343 -349 . DOI: 10.11983/CBB18106
Starch is a main kind of carbohydrates and plays a vital role in energy storage. The granule-bound starch synthase (GBSS) is responsible for the synthesis of amylose. Although GBSS genes have been cloned and identified in many cultivated plants, there are only a few cases of studies on non-cultrivated plants. The present study involved qualitative and quantitative analyses on the sequence characteristics, phylogeny, and expression pattern of GBSS genes in Oryza officinalis. Phylogenetic analysis showed that the GBSS was encoded by two GBSS genes (GBSSI and GBSSII) in all species of Poaceae. In O. officinalis, the two genes shared 62% amino acid identity and displayed different expression patterns in different organs. GBSSII expression was higher in leaves than seeds, whereas GBSSI was mainly expressed in seeds, which suggests divergent spatial expression of the two genes in this wild rice.

| [1] | 包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报 50, 683-690. |
| [2] | 陈凤花, 王琳, 胡丽华 (2005). 实时荧光定量RT-PCR内参基因的选择. 临床检验杂志 23, 393-395. |
| [3] | 顾燕娟 (2006). 支链淀粉合成相关基因等位基因间的差异对稻米淀粉理化特性的影响. 硕士论文. 扬州: 扬州大学. pp. 3-8. |
| [4] | 王倩, 孙文静, 包颖 (2017). 植物颗粒结合淀粉合酶GBSS基因家族的进化. 植物学报 52, 179-187. |
| [5] | 杨学明 (2003). 几个重复序列在不同稻种中的分布及其与稻种分化关系的研究. 硕士论文. 扬州: 扬州大学. pp. 6-10. |
| [6] | 张鹏 (2008). 抑制淀粉分支酶类基因表达对稻米品质影响的研究. 硕士论文. 扬州: 扬州大学. pp. 3-8. |
| [7] | Bao Y, Xu S, Jing X, Meng L, Qin ZY (2015). De novo assembly and characterization ofOryza officinalis leaf transcriptome by using RNA-seq. Biomed Res Int 2015, 982065. |
| [8] | Cheng J, Khan MA, Qiu WM, Li J, Zhou H, Zhang Q, Guo WW, Zhu TT, Peng JH, Sun FJ, Li SH, Korban SS, Han YP (2012). Diversification of genes encoding granulebound starch synthase in monocots and dicots is marked by multiple genome-wide duplication events.PLoS One 7, e30088. |
| [9] | Dian WM, Jiang HW, Wu P (2005). Evolution and expression analysis of starch synthase III and IV in rice.J Exp Bot 56, 623-632. |
| [10] | Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224. |
| [11] | Guindon S, Gascuel O (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Syst Biol 52, 696-704. |
| [12] | Ohdan T, Francisco Jr PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [13] | Shure M, Wessler S, Fedoroff N (1983). Molecular identification and isolation of thewaxy locus in maize. Cell 35, 225-233. |
| [14] | Van Harsselaar JK, Lorenz J, Senning M, Sonnewald U, Sonnewald S (2017). Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genomics 18, 37. |
| [15] | Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264. |
| [16] | Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM (1990). Nucleotide sequence of ricewaxy gene. Nucleic Acids Res 18, 5898. |
/
| 〈 |
|
〉 |