

1西南交通大学生命科学与工程学院, 成都 610031; 2四川省成都市石室中学, 成都 610041
收稿日期: 2025-01-16
修回日期: 2025-04-22
网络出版日期: 2025-05-14
基金资助
国家自然科学基金(No.31900164)、西南交通大学基础培育计划医工结合专项(No.2682023ZTPY073)和西南交通大学SRTP项目(No.2024152, No.2024156)
Expression Pattern and Metabolic Correlation Analysis of TCP Gene Family in Bergenia purpurascens
1College of Life Science and Engineering, Southwest Jiaotong University, Chengdu 610031, China; 2Chengdu Shishi High School of Sichuan Province, Chengdu 610041, China
Received date: 2025-01-16
Revised date: 2025-04-22
Online published: 2025-05-14
TCP蛋白家族是植物特有的一类转录因子家族, 在植物生长发育及响应胁迫过程中发挥着重要作用。为了解岩白菜(Bergenia purpurascens) BpTCP基因家族的功能, 本研究基于转录组测序数据, 利用生物信息学方法对岩白菜BpTCP基因家族进行了系统鉴定与分析。研究共鉴定出16个BpTCP基因, 可分为2大类, 所有BpTCP基因均含有保守的TCP结构域, 相同进化枝的BpTCP蛋白所含基序类型相似。表达模式分析表明, 不同BpTCP基因在不同组织中的表达水平存在差异。在低温胁迫条件下, BpTCP10、BpTCP1和BpTCP12的表达出现显著变化。BpTCP基因家族表达水平与次级代谢物含量的相关性分析显示, 部分BpTCP基因的表达与黄酮类、酚类等多种代谢物的含量之间存在显著相关性。本研究为进一步探究BpTCP基因在岩白菜生长发育、低温胁迫响应及次生代谢合成途径中的生物学功能奠定了基础。
陈靖彧 , 王文庆 , 罗诗语 , 杨路祥 , 汪慧骏 , 吴天宇 , 朱乾坤 . 岩白菜TCP基因家族的表达模式及代谢关联分析[J]. 植物学报, 0 : 1 -0 . DOI: 10.11983/CBB25008
INTRODUCTION: The TCP
protein family is a plant-specific group of transcription factors known to
regulate key biological processes, including growth, development, and stress
responses. Despite their critical roles, the TCP gene family in Bergenia
purpurascens remains uncharacterized. This study aims to systematically
identify and analyze the BpTCP gene family in B. purpurascens using
transcriptome-based bioinformatics approaches, providing insights into their
potential functions in cold adaptation and secondary metabolism.
RATIONALE: B. purpurascens exhibits remarkable resilience to abiotic stresses, particularly cold, and contains abundant secondary metabolites. Given the documented roles of TCP genes in stress responses and metabolic regulation in other plants, we hypothesized that BpTCP genes may contribute to these traits. A comprehensive analysis of this gene family could reveal novel mechanisms underlying stress adaptation and metabolite synthesis, supporting future genetic improvement or biotechnological applications.
RESULTS: Through transcriptome-based bioinformatics analysis, we identified 16 BpTCP genes in B. purpurascens, which were phylogenetically classified into two major groups, with all members containing conserved TCP domains and closely related proteins sharing similar motif patterns. Tissue-specific expression profiling revealed distinct spatial expression patterns across different tissues, suggesting functional diversification among family members. Notably, partial genes, including BpTCP10, BpTCP1 and BpTCP12, exhibited significant expression changes under cold stress, implying their potential cold-responsive roles. Furthermore, expression levels of specific BpTCP genes correlated significantly with accumulation of various secondary metabolites, particularly flavonoids and phenolics, suggesting their regulatory involvement in metabolic pathways.
CONCLUSION: This study provides the first genome-wide characterization of the BpTCP gene family in B. purpurascens, demonstrating its potential roles in growth, cold stress response, and secondary metabolism. The differential expression of BpTCP genes under stress and their correlation with metabolite levels lay a foundation for future functional studies.
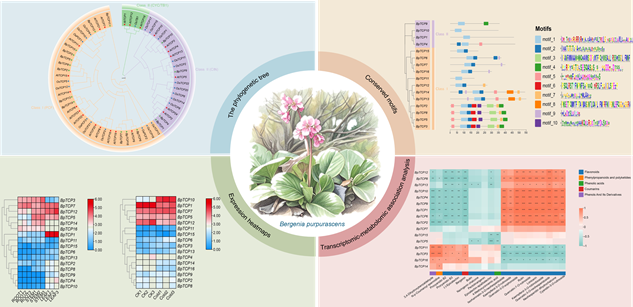
Expression pattern and metabolic correlation analysis of TCP gene family in Bergenia purpurascens. A total of 16 BpTCP genes were identified in Bergenia purpurascens and classified into two major phylogenetic groups. All BpTCP genes contain conserved TCP domains, and proteins from the same evolutionary branch share similar motif compositions. Different BpTCP genes exhibit distinct tissue-specific expression patterns and display distinctive responses to cold stress. Furthermore, certain BpTCP genes demonstrate significant correlations with the accumulation of diverse metabolites.
/
| 〈 |
|
〉 |