

植物学报 ›› 2022, Vol. 57 ›› Issue (4): 490-499.DOI: 10.11983/CBB22080 cstr: 32102.14.CBB22080
黄俊文, 冯琦伊, 郑凯勇, 黄俊杰, 王林博, 赖瑞强, 赖建彬, 阳成伟( )
)
收稿日期:2022-04-12
修回日期:2022-06-23
出版日期:2022-07-01
发布日期:2022-07-14
通讯作者:
阳成伟
作者简介:* E-mail: yangchw@scnu.edu.cn基金资助:Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei
Received:2022-04-12
Revised:2022-06-23
Online:2022-07-01
Published:2022-07-14
About author:First author contact:† These authors contributed equally to this paper
摘要: 蛋白质SUMO化修饰是一种调控蛋白命运的关键修饰方式, 广泛参与植物生长发育及逆境胁迫响应。SUMO化修饰过程主要由激活酶(E1)-结合酶(E2)-连接酶(E3)组成的级联酶促反应催化, 其关键酶组分将SUMO分子缀合至底物蛋白的赖氨酸残基, 形成共价异肽键以完成SUMO化修饰过程。该文报道了1种植物蛋白质SUMO化修饰体外高效检测系统, 通过在大肠杆菌(Escherichia coli)中构建拟南芥(Arabidopsis thaliana) SUMO化修饰的关键通路实现对底物蛋白的SUMO化修饰, 结果可通过免疫印迹进行检测。该系统可以简化植物蛋白质SUMO化修饰的检测流程, 为植物细胞SUMO化修饰的功能研究提供了有力工具。
黄俊文, 冯琦伊, 郑凯勇, 黄俊杰, 王林博, 赖瑞强, 赖建彬, 阳成伟. 植物蛋白质SUMO化修饰体外高效检测系统. 植物学报, 2022, 57(4): 490-499.
Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei. An Effective in Vitro SUMOylation Detection System for Plant Proteins. Chinese Bulletin of Botany, 2022, 57(4): 490-499.
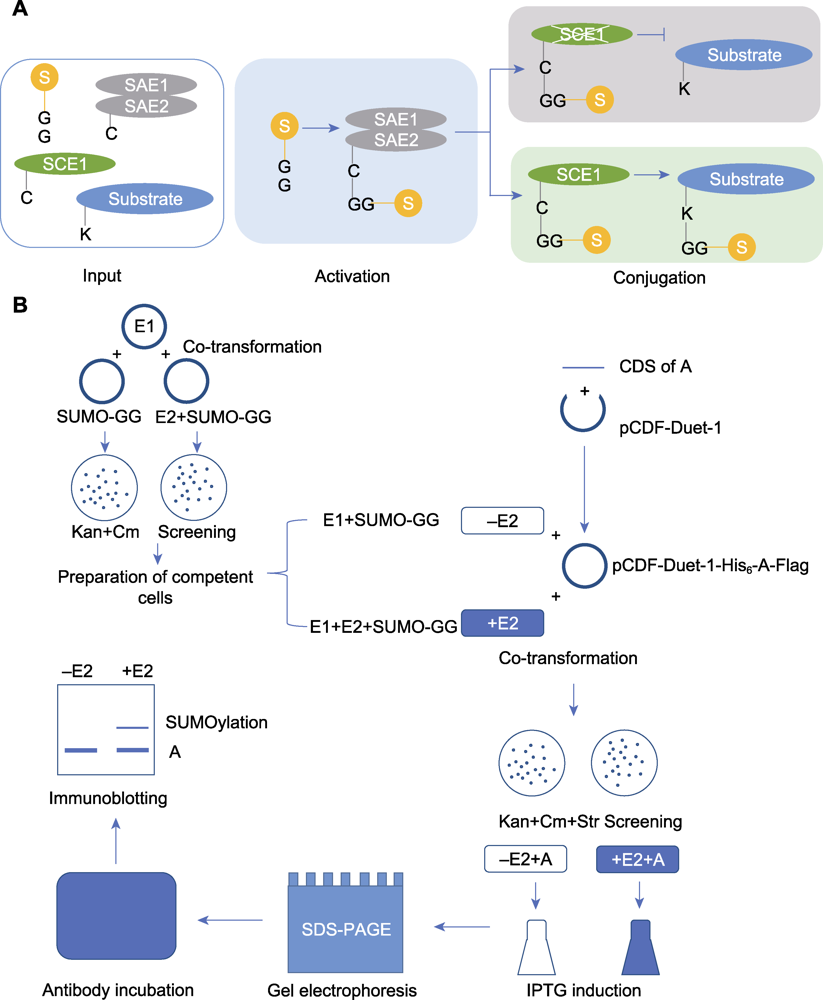
图1 体外SUMO化修饰检测流程 (A) 体外SUMO化修饰检测过程(输入: 将SUMO修饰关键组分基因和候选底物蛋白基因导入大肠杆菌表达; 激活: E1结合以激活SUMO分子参与修饰; 结合: E2将E1激活后的SUMO分子连接至底物完成修饰); (B) 体外SUMO化修饰检测实验流程(共转化: 将SUMO化修饰关键组分载体导入大肠杆菌, 通过卡那霉素和氯霉素平板筛选阳性克隆; 制备感受态细胞: 将获得的阳性克隆制备成感受态细胞; 共转化: 将构建好的底物蛋白表达载体导入感受态细胞, 通过卡那霉素、氯霉素和链霉素平板筛选阳性克隆; 诱导: 通过IPTG诱导蛋白表达; 凝胶电泳: 将获得的菌体样品裂解变性后通过SDS-PAGE凝胶电泳将不同分子量的蛋白分离, 将蛋白从凝胶转移至PVDF膜上; 抗体孵育: 将封闭后的PVDF膜与抗体进行孵育; 显影: 观察并记录实验结果)
Figure 1 The process of in vitro SUMOylation analysis (A) The process diagram of in vitro SUMOylation detection (Input: the key component genes of SUMO modification and the candidate substrate protein gene were introduced into Escherichia coli for expression; Activating: E1 activates SUMO molecules to participate in the modification; Conjugating: E2 attaches the activated SUMO molecules to the substrate); (B) The experimental procedure of in vitro SUMOylation detection (Co-transformation: The plasmids for expressing key SUMOylation components were introduced into E. coli, and the positive colonies were screened on LB agar plates containing kanamycin and chloramphenicol; Preparation of competent cells: The obtained positive colonies were prepared into competent cells; Co-transformation: The constructed plasmid for substrate protein expression was introduced into the competent cells mentioned above, and positive colonies were screened on LB agar plates by kanamycin, chloramphenicol and streptomycin; Induction: Protein expression was induced by IPTG; Gel electrophoresis: After lysed and denatured, samples were separated by SDS-PAGE. Then the proteins were transferred from gel to PVDF membrane; Antibody incubation: The blocked PVDF membrane was incubated with antibody; Immunoblotting: Observation and recording of the experimental results)
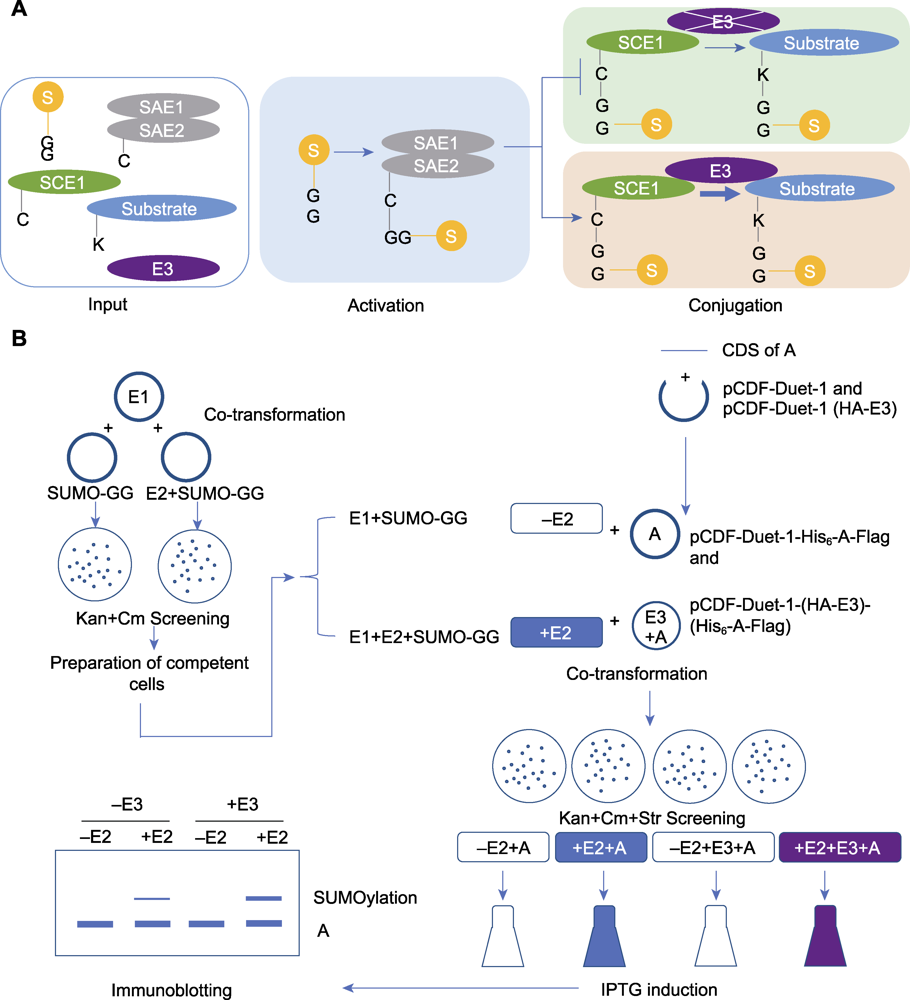
图2 E3连接酶介导的体外SUMO化修饰检测 (A) E3连接酶介导的体外SUMO化修饰检测过程(输入: 将SUMO化修饰关键组分基因和候选底物蛋白基因导入大肠杆菌表达; 激活: E1结合以激活SUMO分子参与修饰; 结合: E2将E1激活后的SUMO分子连接至底物完成修饰, 在E3存在的条件下, E3促进了SUMO分子从E2到底物蛋白的连接); (B) E3连接酶介导的体外SUMO化修饰检测实验流程(共转化: SUMO关键组分载体导入大肠杆菌, 通过卡那霉素和氯霉素平板筛选阳性克隆; 制备感受态细胞: 将获得的阳性克隆制备成感受态细胞; 共转化: 将构建好的底物蛋白表达载体(带有独立的E3表达框)导入感受态细胞, 通过卡那霉素、氯霉素和链霉素平板筛选阳性克隆; 诱导: 通过IPTG诱导蛋白表达; 免疫印迹: 通过SDS-PAGE凝胶电泳将不同分子量的蛋白分离, 将蛋白从凝胶转移至PVDF膜上, 将封闭后的PVDF膜与抗体进行孵育, 通过显影观察并记录实验结果)
Figure 2 The process of in vitro SUMOylation analysis mediated by an E3 ligase (A) The process diagram of the E3 ligase-mediated in vitro SUMOylation detection (Input: The key component genes of SUMO modification and the candidate substrate protein gene were introduced into Escherichia coli for expression; Activating: E1 activates SUMO molecules to participate in the modification; Conjugating: E2 attaches the activated SUMO molecules to the substrate. In the presence of E3, E3 facilitates the attachment of SUMO molecules from E2 to the substrate protein); (B) The experimental procedure of the E3 ligase-mediated in vitro SUMOylation detection (Co-transformation: The plasmids for expression of key SUMOylation components were introduced into E. coli, and the positive colonies were screened on LB agar plates containing kanamycin and chloramphenicol; Preparation of competent cells: The obtained positive colonies were transformed into competent cells; Co-transformation: The constructed plasmid for substrate protein expression (with independent E3 expression cassette) was introduced into the competent cells mentioned above, and the positive colonies were screened by LB agar plates containing kanamycin, chloramphenicol and streptomycin; Induction: Protein expression was induced by IPTG; Immunoblotting: After lysed and denatured, samples were separated by SDS-PAGE gel electrophoresis, then the proteins were transferred from gel to PVDF membrane, the blocked PVDF membrane was incubated with antibody, the result was observed and recorded by immunoblotting).
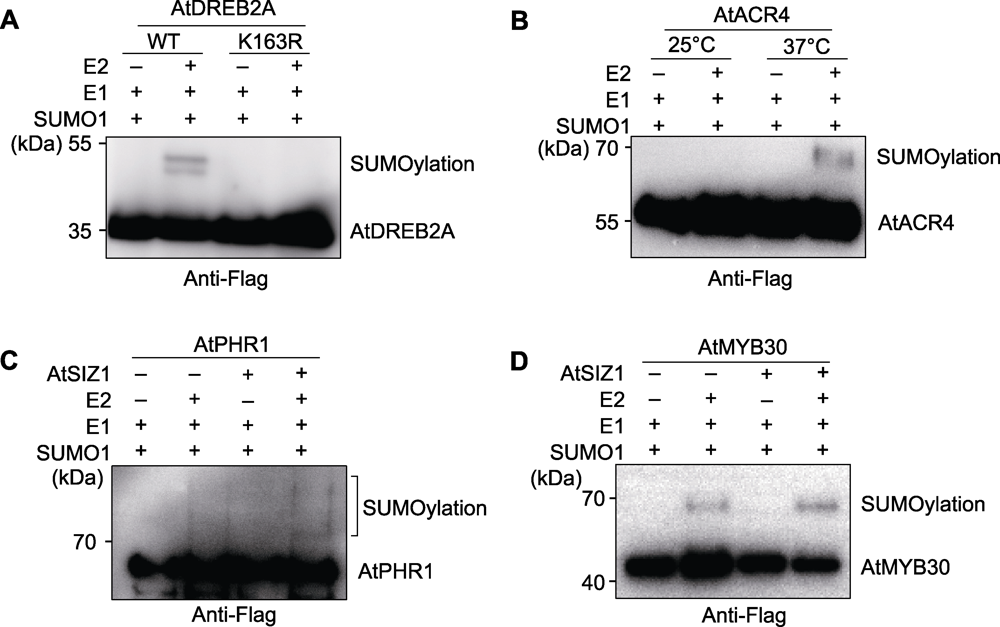
图3 检测实例 (A) AtDREB2A蛋白SUMO化修饰检测; (B) 温度影响AtACR4蛋白的SUMO化修饰水平; (C) E3连接酶AtSIZ1促进AtPHR1蛋白的SUMO化修饰; (D) E3连接酶AtSIZ1促进AtMYB30蛋白的SUMO化修饰
Figure 3 Experimental cases (A) SUMOylation test of AtDREB2A; (B) Temperature affects SUMOylation of AtACR4; (C) The E3 ligase AtSIZ1 facilitates SUMOylation of AtPHR1; (D) The E3 ligase AtSIZ1 facilitates SUMOylation of AtMYB30
| [1] |
韩丹璐, 赖建彬, 阳成伟 (2018). SUMO E3连接酶在植物生长发育中的功能研究进展. 植物学报 53, 175-184.
DOI |
| [2] |
曲高平, 金京波 (2020). 植物蛋白SUMO化修饰检测方法. 植物学报 55, 83-89.
DOI |
| [3] |
Augustine RC, Vierstra RD (2018). SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol 45, 143-154.
DOI PMID |
| [4] | Dai Vu L, Gevaert K, De Smet I (2018). Protein language: post-translational modifications talking to each other. Tren- ds Plant Sci 23, 1068-1080. |
| [5] |
Fang Q, Zhang J, Zhang Y, Fan N, van den Burg HA, Huang CF (2020). Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32, 3921-3938.
DOI URL |
| [6] |
Han DL, Lai JB, Yang CW (2021). SUMOylation: a critical transcription modulator in plant cells. Plant Sci 310, 110987.
DOI URL |
| [7] |
Huang LX, Yang SG, Zhang SC, Liu M, Lai JB, Qi YL, Shi SF, Wang JX, Wang YQ, Xie Q, Yang CW (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60, 666-678.
DOI URL |
| [8] |
Jiang JM, Xie Y, Du JJ, Yang CW, Lai JB (2021). A SUMO ligase OsMMS21 regulates rice development and auxin response. J Plant Physiol 263, 153447.
DOI URL |
| [9] |
Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53, 530-540.
DOI URL |
| [10] |
Kong XF, Hong YC, Hsu YF, Huang H, Liu X, Song Z, Zhu JK (2020). SIZ1-mediated SUMOylation of ROS1 enhances its stability and positively regulates active DNA demethylation in Arabidopsis. Mol Plant 13, 1816-1824.
DOI URL |
| [11] |
Li YR, Williams B, Dickman M (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)- mediated heat tolerance requires translocation, SUMOylation and binding to WRKY29. New Phytol 214, 695-705.
DOI URL |
| [12] |
Liu YY, Lai JB, Yu MY, Wang FG, Zhang JJ, Jiang JM, Hu H, Wu Q, Lu GH, Xu PL, Yang CW (2016). The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/ DPa complex in cell cycle regulation. Plant Cell 28, 2225-2237.
DOI URL |
| [13] |
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007). SIZ1-mediated SUMOylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403-1414.
DOI URL |
| [14] |
Miura K, Lee J, Miura T, Hasegawa PM (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51, 103-113.
DOI URL |
| [15] |
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102, 7760-7765.
DOI URL |
| [16] |
Morrell R, Sadanandom A (2019). Dealing with stress: a review of plant SUMO proteases. Front Plant Sci 10, 1122.
DOI URL |
| [17] |
Niu D, Lin XL, Kong XX, Qu GP, Cai B, Lee J, Jin JB (2019). SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12, 215-228.
DOI PMID |
| [18] |
Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama JI, Tanaka K (2009). Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50, 1049-1061.
DOI URL |
| [19] |
Perry JJP, Tainer JA, Boddy MN (2008). A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33, 201-208.
DOI URL |
| [20] |
Roy D, Sadanandom A (2021). SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants. Cell Mol Life Sci 78, 2641-2664.
DOI URL |
| [21] |
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress- responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103, 18822-18827.
DOI URL |
| [22] |
Saleh A, Withers J, Mohan R, Marqués J, Gu YN, Yan SP, Zavaliev R, Nomoto M, Tada Y, Dong XN (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169-182.
DOI URL |
| [23] |
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145, 119-134.
PMID |
| [24] | Verma V, Srivastava AK, Gough C, Campanaro A, Srivastava M, Morrell R, Joyce J, Bailey M, Zhang CJ, Krysan PJ, Sadanandom A (2021). SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc Natl Acad Sci USA 118, e2021351118. |
| [25] | Wang FG, Liu YY, Shi YQ, Han DL, Wu YY, Ye WX, Yang HL, Li GW, Cui F, Wan SB, Lai JB, Yang CW (2020). SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant Physiol 183, 41-50. |
| [26] |
Wang Z, Prelich G (2009). Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 29, 1694-1706.
DOI URL |
| [27] |
Xu JM, Zhu JY, Liu JJ, Wang JX, Ding ZJ, Tian HY (2021). SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis. J Integr Plant Biol 63, 1147-1160.
DOI URL |
| [28] |
Zhang JJ, Lai JB, Wang FG, Yang SG, He ZP, Jiang JM, Li QL, Wu Q, Liu YY, Yu MY, Du JJ, Xie Q, Wu KQ, Yang CW (2017). A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant Physiol 173, 1574-1582.
DOI URL |
| [29] |
Zheng Y, Schumaker KS, Guo Y (2012). SUMOylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109, 12822-12827.
DOI URL |
| [30] |
Zhou LJ, Zhang CL, Zhang RF, Wang GL, Li YY, Hao YJ (2019). The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis. Plant Physiol 179, 88-106.
DOI PMID |
| [1] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [2] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [3] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [4] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [5] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [14] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [15] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||