

植物学报 ›› 2020, Vol. 55 ›› Issue (4): 457-467.DOI: 10.11983/CBB19228 cstr: 32102.14.CBB19228
收稿日期:2019-11-23
接受日期:2020-03-01
出版日期:2020-07-01
发布日期:2020-05-21
通讯作者:
姚纲
基金资助:
Jiuxiang Huang1,Wenna Chen2,Yuling Li1,Gang Yao1,*( )
)
Received:2019-11-23
Accepted:2020-03-01
Online:2020-07-01
Published:2020-05-21
Contact:
Gang Yao
摘要: 苋科(Amaranthaceae sensu lato)是石竹目(Caryophyllales)第二大科, 目前被普遍接受的苋科为其广义概念, 含狭义苋科(Amaranthaceae sensu stricto)和藜科(Chenopodiaceae)。然而到目前为止, 藜科是否应作为独立的科还存在争议。此外, 广义苋科内部各亚科之间的系统关系也尚未厘清。对广义苋科所有13个亚科代表类群进行取样(共59种), 基于8个叶绿体序列片段重建其系统发育关系, 并结合分子钟估算, 对该科及其主要分支的起源与分化时间进行推测。结果表明, 广义苋科与狭义苋科都是很好的单系, 但藜科并非单系, 因此不支持藜科在科级水平的地位, 支持广义苋科的观点。除了多节草亚科(Polycnemoideae)之外, 其它亚科的系统位置均得到很好的分辨。分子钟估算结果表明, 广义苋科于白垩纪晚期约69.9 Ma分化出该科的2个主要分支, 且该科在白垩纪-古近纪边界附近时期(约66.0 Ma)可能发生过快速辐射分化事件。
黄久香,陈文娜,李玉玲,姚纲. 基于多个叶绿体基因序列片段重建广义苋科系统发育关系. 植物学报, 2020, 55(4): 457-467.
Jiuxiang Huang,Wenna Chen,Yuling Li,Gang Yao. Phylogenetic Study of Amaranthaceae sensu lato Based on Multiple Plastid DNA Fragments. Chinese Bulletin of Botany, 2020, 55(4): 457-467.
| Name of DNA sequences | Length of DNA sequences (bp) | Model selected |
|---|---|---|
| atpB | 1497 | GTR+I+Γ |
| ndhF | 2242 | TVM+I+Γ |
| psbB | 1527 | GTR+I+Γ |
| rbcL | 1344 | TVM+I+Γ |
| rpoC2 | 4147 | GTR+I+Γ |
| rps4 | 606 | TVM+Γ |
| rps16 | 1401 | GTR+I+Γ |
| matK/trnK | 2757 | GTR+I+Γ |
表1 本研究所用DNA序列相关信息
Table 1 Information of DNA sequences used in the study
| Name of DNA sequences | Length of DNA sequences (bp) | Model selected |
|---|---|---|
| atpB | 1497 | GTR+I+Γ |
| ndhF | 2242 | TVM+I+Γ |
| psbB | 1527 | GTR+I+Γ |
| rbcL | 1344 | TVM+I+Γ |
| rpoC2 | 4147 | GTR+I+Γ |
| rps4 | 606 | TVM+Γ |
| rps16 | 1401 | GTR+I+Γ |
| matK/trnK | 2757 | GTR+I+Γ |
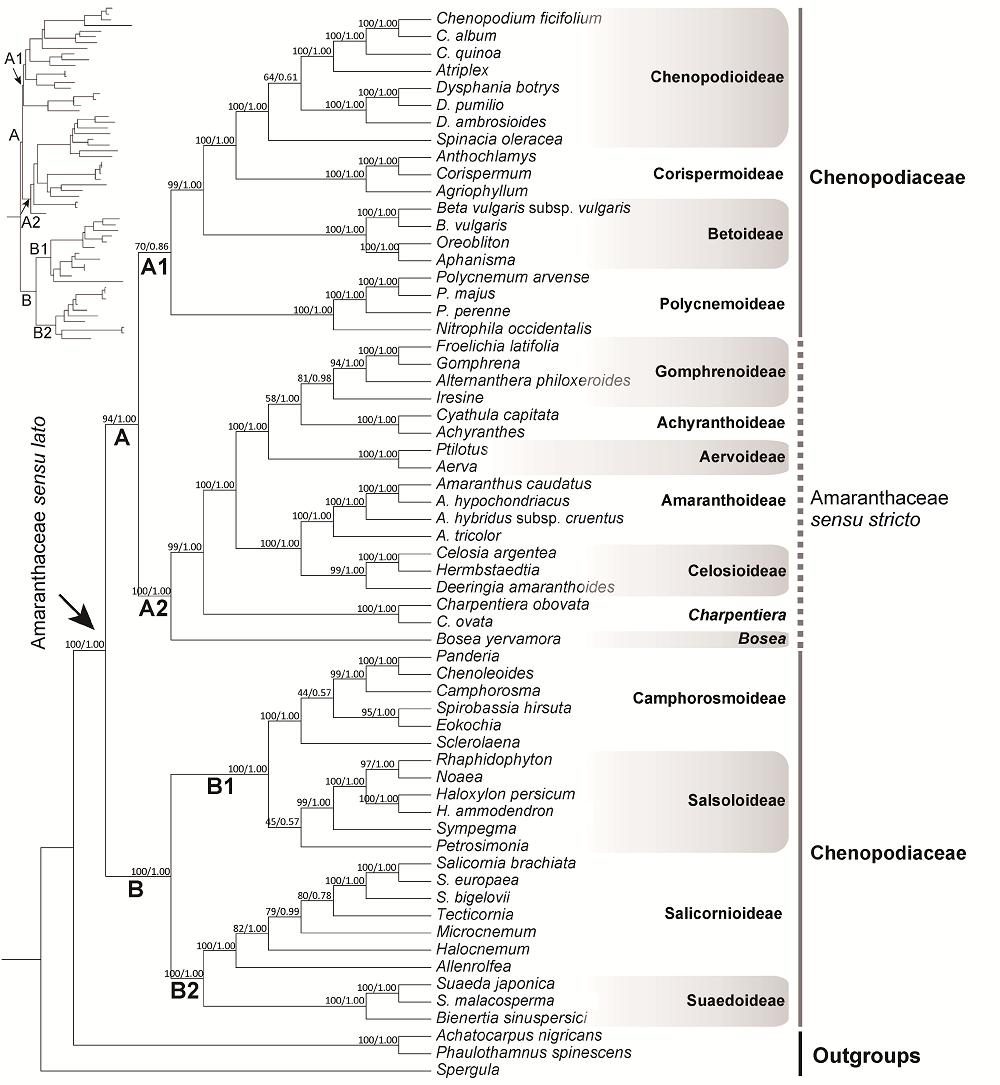
图1 基于8个叶绿体DNA序列片段构建的广义苋科系统发育树 各分支节点上方数字分别代表ML树自展支持率/BI树后验概率。
Figure 1 Phylogeny of Amaranthaceae sensu lato inferred from the combined matrix of eight plastid DNA regions Bootstrap support value and posterior probability of each node are indicated above branches.
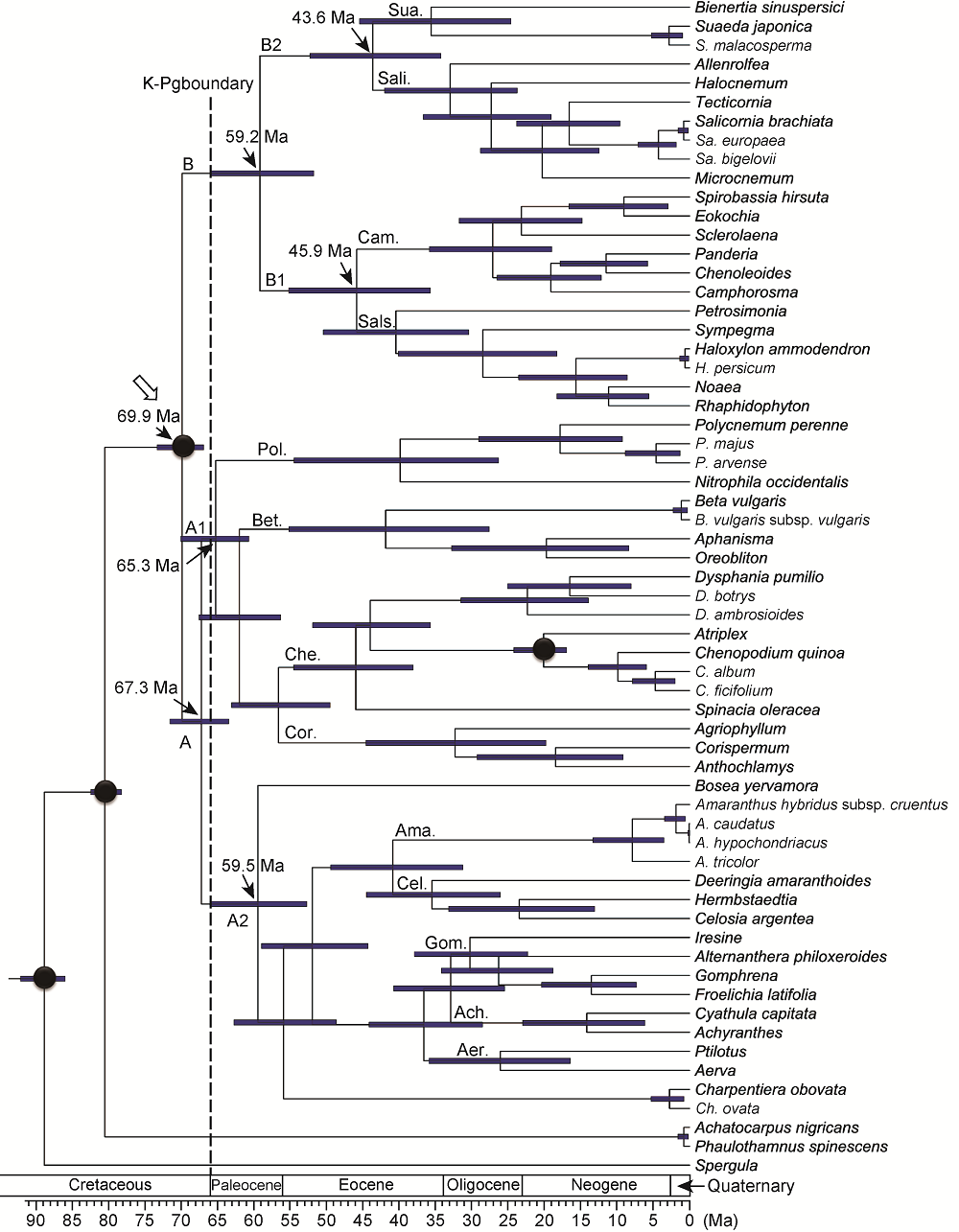
图2 基于8个叶绿体DNA序列片段及4个时间键准点采用BEAST分析所得广义苋科分化时间 黑色圆点表示时间键准点所在位置。空心箭头示广义苋科冠群节点。Ach: 牛膝亚科; Aer: 白花苋亚科; Ama: 苋亚科; Bet: 甜菜亚科; Cam: 樟味藜亚科; Cel: 青葙亚科; Che: 藜亚科; Cor: 虫实亚科; Gom: 千日红亚科; Pol: 多节草亚科; Sali: 盐角草亚科; Sals: 猪毛菜亚科; Sua: 碱蓬亚科
Figure 2 Chronogram based on the BEAST analysis of the combined matrix of eight plastid DNA regions and four calibrations Calibration points are depicted with black circles. The crown node of Amaranthaceae sensu lato is shown by a hollow arrowhead. Ach: Achyranthoideae; Aer: Aervoideae; Ama: Amaranthoideae; Bet: Betoideae; Cam: Camphorosmoideae; Cel: Celosioideae; Che: Chenopodioideae; Cor: Corispermoideae; Gom: Gomphrenoideae; Pol: Polycnemoideae; Sali: Salicornioideae; Sals: Salsoloideae; Sua: Suaedoideae
| [1] | 中国科学院中国植物志编辑委员会 (1979). 中国植物志, 第25卷第2分册. 北京: 科学出版社. pp. 241. |
| [2] | APG IV (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181, 1-20. |
| [3] | Bentham G, Hooker JD (1880). Genera Plantarum, Vol. 3. London: Lovell Reeve. pp. 1258. |
| [4] | Brockington SF, Alexandre R, Ramdial J, Moore MJ, Crawley S, Dhingra A, Hilu K, Soltis DE, Soltis PS (2009). Phylogeny of the Caryophyllales sensu lato: revisiting hypotheses on pollination biology and perianth differentiation in the core Caryophyllales. Int J Plant Sci 170, 627-643. |
| [5] | Christenhusz MJM, Byng JW (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201-217. |
| [6] | Cronquist A (1988). Caryophyllidae. In: The Evolution and Classification of Flowering Plants, 2nd edn. New York: The New York Botanical Garden. pp. 309-320. |
| [7] | Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002). Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89, 132-144. |
| [8] | Drummond AJ, Suchard MA, Xie D, Rambaut A (2012). Bayesian phylogenetics with BEAUTi and the BEAST 1.7. Mol Biol Evol 29, 1967-1973. |
| [9] | Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, Endara L, Kriebel R, Neubig KM, Whitten WM, Williams NH, Cameron KM (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B Biol Sci 282, 20151553. |
| [10] | Guo X, Thomas DC, Saunders RMK (2018). Gene tree discordance and coalescent methods support ancient intergeneric hybridisation between Dasymaschalon and Friesodielsia(Annonaceae). Mol Phylogenet Evol 127, 14-29. |
| [11] | Gurushidze M, Fritsch RM, Blattner FR (2010). Species-level phylogeny of Allium subgenus Melanocrommyum: incomplete lineage sorting, hybridization and trnF gene duplication. Taxon 59, 829-840. |
| [12] | Hernández-Ledesma P, Berendsohn WG, Borsch T, von Mering S, Akhani H, Arias S, Castañeda-Noa I, Eggli U, Eriksson R, Flores-Olvera H, Fuentes-Bazán S, Kadereit G, Klak C, Korotkova N, Nyffeler R, Ocampo G, Ochoterena H, Oxelman B, Rabeler RK, Sanchez A, Schlumpberger BO, Uotila P (2015). A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia 45, 281-383. |
| [13] | Kadereit G, Ackerly D, Pirie MD (2012). A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc R Soc B Biol Sci 279, 3304-3311. |
| [14] |
Kadereit G, Borsch T, Weising K, Freitag H (2003). Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int J Plant Sci 164, 959-986.
DOI URL |
| [15] | Kadereit G, Gotzek D, Jacobs S, Freitag H (2005). Origin and age of Australian Chenopodiaceae. Org Divers Evol 5, 59-80. |
| [16] |
Katoh K, Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772-780.
URL PMID |
| [17] | Koenen EJM, Clarkson JJ, Pennington TD, Chatrou LW (2015). Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol 207, 327-339. |
| [18] | Kühn U, Bittrich V, Carolin R, Freitag H, Hedge IC, Uotila P, Wilson PG (1993). Chenopodiaceae. In: Kubitzki K, ed. Families and Genera of Vascular Plants, Vol. 2. Berlin: Springer. pp. 253-281. |
| [19] |
Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants 5, 461-470.
URL PMID |
| [20] |
Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015). A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207, 437-453.
URL PMID |
| [21] | Masson R, Kadereit G (2013). Phylogeny of Polycnemoideae (Amaranthaceae): implications for biogeography, character evolution and taxonomy. Taxon 62, 100-111. |
| [22] | Miller MA, Pfeiffer W, Schwartz T (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA: IEEE. pp. 1-8. |
| [23] | Moquin-Tandon A (1837). Note sur le genre Polycnemum et sur une nouvelle tribu de la famille des Paronychees. Ann Sci Nat 7, 33-42. |
| [24] | Muller J (1981). Fossil pollen records of extant angiosperms. Bot Rev 47, 1-142. |
| [25] | Müler K, Borsch T (2005). Phylogenetics of Amaranthaceae based on matK/trnK sequence data—evidence from parsimony, likelihood, and Bayesian analyses. Ann Missouri Bot Gard 92, 66-102. |
| [26] |
Posada D (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253-1256.
DOI URL PMID |
| [27] | Pratt DB (2003). Phylogeny and Morphological Evolution of the Chenopodiaceae-Amaranthaceae Alliance. Ph.D. thesis. Ames: Iowa State University. pp. 116. |
| [28] | Rambaut A (2012). FigTree version 1.4.0. Available from: http://tree.bio.ed.ac.uk/software/figtree/. |
| [29] | Rambaut A, Suchard MA, Drummond AJ (2014). Tracer v1.6. Available from: http://beast.bio.ed.ac.uk/Tracer/. |
| [30] |
Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574.
URL PMID |
| [31] |
Schulte P, Alegret L, Arenillas I, Arz JA, Barton PJ, Bown PR, Bralower TJ, Christeson GL, Claeys P, Cockell CS, Collins GS, Deutsch A, Goldin TJ, Goto K, Grajales-Nishimura JM, Grieve RAF, Gulick SPS, Johnson KR, Kiessling W, Koeberl C, Kring DA, MacLeod KG, Matsui T, Melosh J, Montanari A, Morgan JV, Neal CR, Nichols DJ, Norris RD, Pierazzo E, Ravizza G, Rebolledo-Vieyra M, Reimold WU, Robin E, Salge T, Speijer RP, Sweet AR, Urrutia-Fucugauchi J, Vajda V, Whalen MT, Willumsen PS (2010). The chicxulub asteroid impact and mass extinction at the Cretaceous-Paleogene boundary. Science 327, 1214-1218.
DOI URL PMID |
| [32] | Scott AJ (1977a). Nomina conservanda proposita. Taxon 26, 246. |
| [33] | Scott AJ (1977b). Reinstatement and revision of Salicorniaceae J. Agardh (Caryophyllales). Bot J Linn Soc 75, 357-374. |
| [34] |
Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, Bell CD, Latvis M, Crawley S, Black C, Diouf D, Xi ZX, Rushworth CA, Gitzendanner MA, Sytsma KJ, Qiu YL, Hilu KW, Davis CC, Sanderson MJ, Beaman RS, Olmstead RG, Judd WS, Donoghue MJ, Soltis PS (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot 98, 704-730.
URL PMID |
| [35] | Soriano A (1944). El género Nitrophila en la Argentina y su posiciónsistemática. Rev Argent Agron 11, 302. |
| [36] |
Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690.
DOI URL PMID |
| [37] | Stevens PF (2001). Angiosperm Phylogeny Website. Version 14, July 2017 [and more or less continuously updated since]. Available from: http://www.mobot.org/MOBOT/research/APweb/. |
| [38] |
Sukhorukov AP, Mavrodiev EV, Struwig M, Nilova MV, Dzhalilova KK, Balandin SA, Erst A, Krinitsyna AA (2015). One-seeded fruits in the core Caryophyllales: their origin and structural diversity. PLoS One 10, e0117974.
URL PMID |
| [39] | Takhtajan A (1997). Diversity and Classification of Flowering Plants. New York: Columbia University Press. pp. 1-643. |
| [40] | Ulbrich E (1934). Chenopodiaceae. In: Engler A, Prantl K, eds. Die Naturlichen Pflanzenfamilien, Vol. 16c. Leipzig: Engelmann. pp. 379-584. |
| [41] |
Walker JF, Yang Y, Feng T, Timoneda A, Mikenas J, Hutchison V, Edwards C, Wang N, Ahluwalia S, Olivieri J, Walker-Hale N, Majure LC, Puente R, Kadereit G, Lauterbach M, Eggli U, Flores-Olvera H, Ochoterena H, Brockington SF, Moore MJ, Smith SA (2018). From cacti to carnivores: improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. Am J Bot 105, 446-462.
DOI URL PMID |
| [42] | Wang W, Ortiz RDC, Jacques FMB, Xiang XG, Li HL, Lin L, Li RQ, Liu Y, Soltis PS, Soltis DE, Chen ZD (2012). Menispermaceae and the diversification of tropical rainforests near the Cretaceous-Paleogene boundary. New Phytol 195, 470-478. |
| [43] | Wikström N, Savolainen V, Chase MW (2001). Evolution of the angiosperms: calibrating the family tree. Proc R Soc B Biol Sci 268, 2211-2220. |
| [44] |
Yang Y, Moore MJ, Brockington SF, Mikenas J, Olivieri J, Walker JF, Smith SA (2018). Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytol 217, 855-870.
DOI URL PMID |
| [45] |
Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, Soltis DE, Soltis PS, Smith SA, Brockington SF, Moore MJ, Yi TS, Li DZ (2019). Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol 134, 74-86.
DOI URL PMID |
| [46] |
Yi TS, Jin GH, Wen J (2015). Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian—New World disjunct plant genus Osmorhiza(Apiaceae). Mol Phylogenet Evol 85, 10-21.
URL PMID |
| [1] | 孙亚君. 何谓高等或低等生物——澄清《物种起源》所蕴含的生物等级性的涵义及其成立性[J]. 生物多样性, 2025, 33(1): 24394-. |
| [2] | 艾妍雨, 胡海霞, 沈婷, 莫雨轩, 杞金华, 宋亮. 附生维管植物多样性及其与宿主特征的相关性: 以哀牢山中山湿性常绿阔叶林为例[J]. 生物多样性, 2024, 32(5): 24072-. |
| [3] | 陈文娜, 李良涛, 周璐, 姚纲. 太行山近期隆升促进太行花属(蔷薇科)谱系分化[J]. 植物学报, 2024, 59(5): 763-773. |
| [4] | 吕燕文, 王子韵, 肖钰, 何梓晗, 吴超, 胡新生. 谱系分选理论与检测方法的研究进展[J]. 生物多样性, 2024, 32(4): 23400-. |
| [5] | 曹可欣, 王敬雯, 郑国, 武鹏峰, 李英滨, 崔淑艳. 降水格局改变及氮沉降对北方典型草原土壤线虫多样性的影响[J]. 生物多样性, 2024, 32(3): 23491-. |
| [6] | 王斌, 钟艺倩, 杨美雪, 吴淼锐, 王艳萍, 陆芳, 陶旺兰, 李健星, 赵弘明, 刘晟源, 向悟生, 李先琨. 喀斯特季节性雨林优势树种叶片非结构性碳水化合物空间变异及生态驱动因素[J]. 生物多样性, 2024, 32(12): 24325-. |
| [7] | 何林君, 杨文静, 石宇豪, 阿说克者莫, 范钰, 王国严, 李景吉, 石松林, 易桂花, 彭培好. 火烧干扰下植物群落系统发育和功能多样性对紫茎泽兰入侵的影响[J]. 生物多样性, 2024, 32(11): 24269-. |
| [8] | 杨向林, 赵彩云, 李俊生, 种方方, 李文金. 植物入侵导致群落谱系结构更加聚集: 以广西国家级自然保护区草本植物为例[J]. 生物多样性, 2024, 32(11): 24175-. |
| [9] | 王振宇, 黄志群. 亚热带27种木本植物叶片性状对植食作用的影响: 验证生长-防御权衡假说[J]. 植物生态学报, 2024, 48(11): 1501-1509. |
| [10] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [11] | 褚振州, 古丽巴哈尔·依斯拉木, 屈泽众, 田新民. 同域分布的3种木蓼属植物叶绿体基因组比较[J]. 植物学报, 2023, 58(3): 417-432. |
| [12] | 宋会银, 胡征宇, 刘国祥. 绿藻门小球藻科的分类学研究进展[J]. 生物多样性, 2023, 31(2): 22083-. |
| [13] | 李治中, 彭帅, 王青锋, 李伟, 梁士楚, 陈进明. 中国海菜花属植物隐种多样性[J]. 生物多样性, 2023, 31(2): 22394-. |
| [14] | 包金波, 丁志杰, 苗浩宇, 李雪丽, 任书贤, 焦若岩, 李浩, 邓茜茜, 李英姿, 田新民. 石栗叶绿体基因组研究[J]. 植物学报, 2023, 58(2): 248-260. |
| [15] | 朱瑞良, 马晓英, 曹畅, 曹子寅. 中国苔藓植物多样性研究进展[J]. 生物多样性, 2022, 30(7): 22378-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||