

植物学报 ›› 2024, Vol. 59 ›› Issue (5): 774-782.DOI: 10.11983/CBB24107 cstr: 32102.14.CBB24107
收稿日期:2024-07-18
接受日期:2024-08-20
出版日期:2024-09-10
发布日期:2024-08-22
通讯作者:
陈春丽
基金资助:
Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen*( )
)
Received:2024-07-18
Accepted:2024-08-20
Online:2024-09-10
Published:2024-08-22
Contact:
Chunli Chen
摘要: 流式细胞术是一种高通量技术, 可以同时快速检测单个颗粒物的多项物理及生物学特征。随着测序成本的大幅降低, 流式细胞术在植物基因组学高通量样品获取中的作用日益凸显。该文以水稻(Oryza sativa)和大豆(Glycine max)为例, 详细介绍了应用流式细胞术对植物细胞核进行精细分选以及后续的ATAC-seq和RNA-seq实验分析流程, 为农业生物育种中基因的高效挖掘提供了优选工具。同时针对实验操作中的关键技术和常见问题, 如细胞核制备注意事项、分选纯度与效率的平衡及单细胞分选调试方法进行分析并提出建议, 为植物科学工作者应用流式细胞术开展基因组学研究提供参考。
夏春皎, 李运广, 夏姝, 庞伟, 陈春丽. 植物基因组学中的流式细胞分析及分选技术. 植物学报, 2024, 59(5): 774-782.
Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics. Chinese Bulletin of Botany, 2024, 59(5): 774-782.
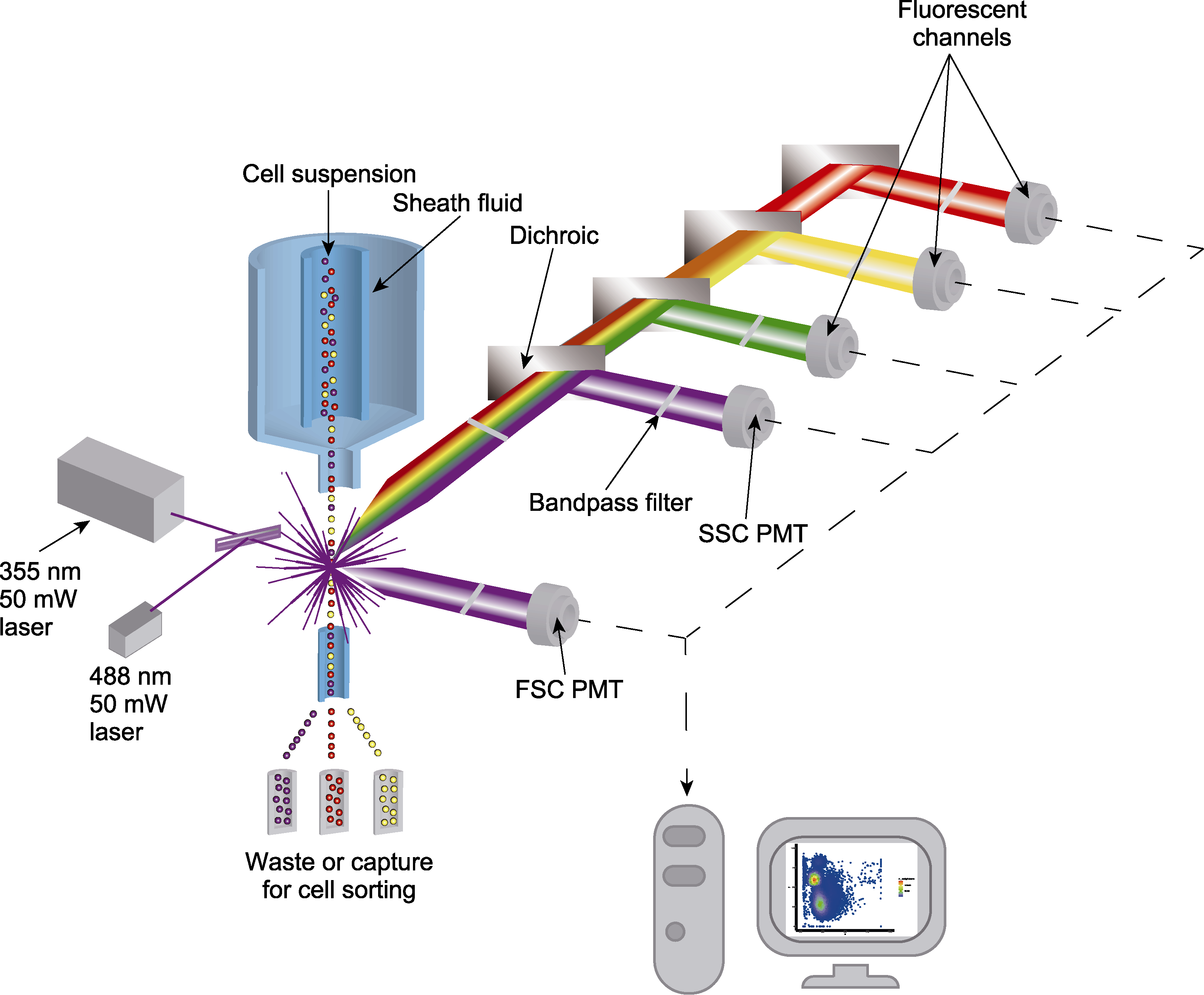
图1 流式细胞仪的基本结构 SSC: 侧向散射通道; FSC: 前向散射通道; PMT: 光电倍增管
Figure 1 Basic structure of flow cytometer SSC: Side scatter channel; FSC: Forward scatter channel; PMT: Photomultiplier tube
| Names | Components | Applications |
|---|---|---|
| Galbraith’s (Galbraith et al., | 45 mmol∙L-1 MgCl2, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 0.1% (v/v) TritonX-100, pH7.0 | Arabidopsis thaliana, Glycine max |
| LB01 (Dpooležel et al., | 15 mmol∙L-1 Tris, 2 mmol∙L-1 Na2EDTA, 0.5 mmol∙L-1 spermine, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.1% (v/v) TritonX-100, 15 mmol∙L-1 β-mercaptoethanol, pH7.0-8.0 | Oryza sativa, Chrysanthemum indicum, Solanum lycopersicum |
| Otto’s (Otto, | OTTO I: 100 mmol∙L-1 citric acid, 0.5% (v/v) Tween-20, pH2.0- 3.0; OTTO II: 400 mmol∙L-1 Na2HPO4·12H2O, pH8.0-9.0 | Ranunculus japonicus, O. sativa |
| Tris-MgCl2 (Pfosser et al., | 200 mmol∙L-1 Tris, 4 mmol∙L-1 MgCl2·6H2O, 0.5% (v/v) TritonX- 100, pH7.5 | Centaurea cyanus, Celtis au- stralis |
| GPB (Loureiro et al., | 0.5 mmol∙L-1 spermine, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.5% (v/v) TritonX- 100, pH7.0 | O. sativa, Actinidia chinensis |
| WPB (Loureiro et al., | 0.2 mol∙L-1 Tris-HCl, 4 mmol∙L-1 MgCl2∙6H2O, 2 mmol∙L-1 EDTA Na2·2H2O, 86 mmol∙L-1 NaCl, 10 mmol∙L-1 sodium pyrosulfite, 1% PVP-10, 1% (v/v) TritonX-100, pH7.5 | Vitis vinifera, Quercus robur |
| PVPK12-mGB2 (Zhang and Feng, | 30 mmol∙L-1 sodium citrate, 45 mmol∙L-1 MgCl2, 20 mmol∙L-1 MO- PS, 20 mmol∙L-1 NaCl, 20 mmol∙L-1 EDTA Na2·2H2O, 0.1% (v/v) TritonX-100, 0.5% (v/v) Tween-20, 10 μL·mL-1 β-mercaptoethanol, 1%-2% PVPK12, pH7.0 | Silicone fast-drying plant ma- terials |
表1 几种代表性的植物细胞核解离缓冲液
Table 1 Several representative plant cell nucleus dissociation buffers
| Names | Components | Applications |
|---|---|---|
| Galbraith’s (Galbraith et al., | 45 mmol∙L-1 MgCl2, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 0.1% (v/v) TritonX-100, pH7.0 | Arabidopsis thaliana, Glycine max |
| LB01 (Dpooležel et al., | 15 mmol∙L-1 Tris, 2 mmol∙L-1 Na2EDTA, 0.5 mmol∙L-1 spermine, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.1% (v/v) TritonX-100, 15 mmol∙L-1 β-mercaptoethanol, pH7.0-8.0 | Oryza sativa, Chrysanthemum indicum, Solanum lycopersicum |
| Otto’s (Otto, | OTTO I: 100 mmol∙L-1 citric acid, 0.5% (v/v) Tween-20, pH2.0- 3.0; OTTO II: 400 mmol∙L-1 Na2HPO4·12H2O, pH8.0-9.0 | Ranunculus japonicus, O. sativa |
| Tris-MgCl2 (Pfosser et al., | 200 mmol∙L-1 Tris, 4 mmol∙L-1 MgCl2·6H2O, 0.5% (v/v) TritonX- 100, pH7.5 | Centaurea cyanus, Celtis au- stralis |
| GPB (Loureiro et al., | 0.5 mmol∙L-1 spermine, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.5% (v/v) TritonX- 100, pH7.0 | O. sativa, Actinidia chinensis |
| WPB (Loureiro et al., | 0.2 mol∙L-1 Tris-HCl, 4 mmol∙L-1 MgCl2∙6H2O, 2 mmol∙L-1 EDTA Na2·2H2O, 86 mmol∙L-1 NaCl, 10 mmol∙L-1 sodium pyrosulfite, 1% PVP-10, 1% (v/v) TritonX-100, pH7.5 | Vitis vinifera, Quercus robur |
| PVPK12-mGB2 (Zhang and Feng, | 30 mmol∙L-1 sodium citrate, 45 mmol∙L-1 MgCl2, 20 mmol∙L-1 MO- PS, 20 mmol∙L-1 NaCl, 20 mmol∙L-1 EDTA Na2·2H2O, 0.1% (v/v) TritonX-100, 0.5% (v/v) Tween-20, 10 μL·mL-1 β-mercaptoethanol, 1%-2% PVPK12, pH7.0 | Silicone fast-drying plant ma- terials |
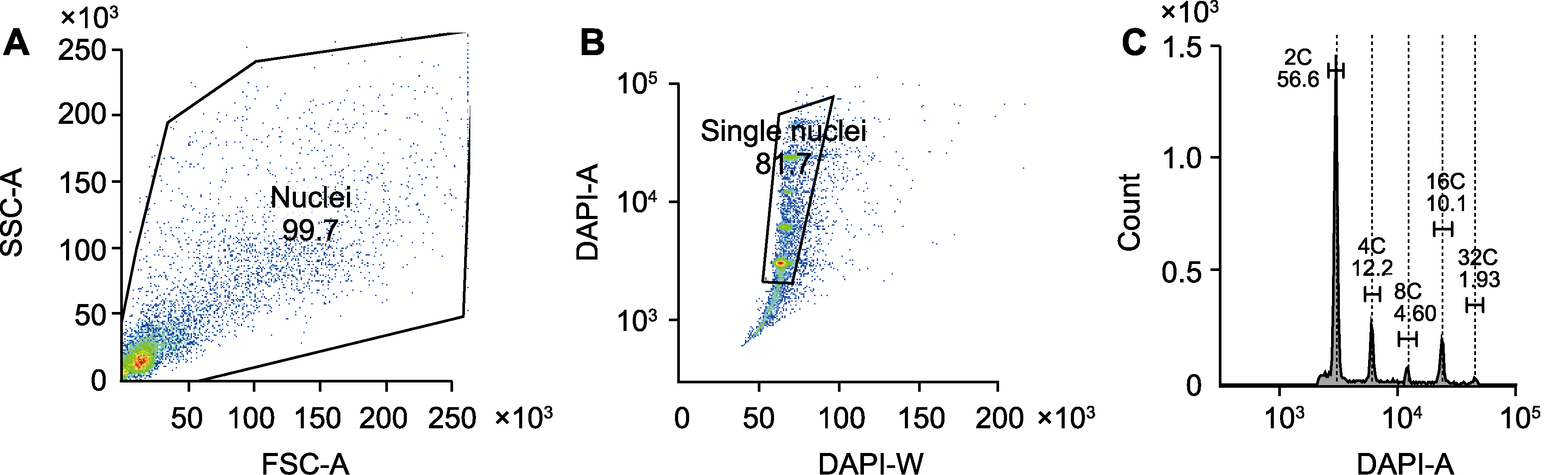
图2 大豆根瘤细胞核不同DNA倍性分析 (A) 通过FSC-A与SSC-A双参数设定总细胞核门; (B) 通过DAPI-W和DAPI-A设定单细胞核门; (C) 通过DAPI-A直方图设定不同DNA倍性的细胞核门。SSC和FSC同图1。DAPI: 4,6-二脒基-2-苯基吲哚
Figure 2 Analysis of different DNA ploidies in nuclei of soybean nodule cells (A) Total nucleus gate was set by FSC-A and SSC-A dual parameters; (B) Single nuclei gate was set by DAPI-W and DAPI-A; (C) Nuclei gate for different DNA ploidies was set by DAPI-A histogram. SSC and FSC are the same as shown in Figure 1. DAPI: 4,6-diamidino-2-phenylindole
| [1] | Bressan D, Battistoni G, Hannon GJ (2023). The dawn of spatial omics. Science 381, eabq4964. |
| [2] | Büscher M (2019). Flow cytometry instrumentation—an overview. Curr Protoc Cytom doi: 10.1002/cpcy.52. |
| [3] | Cápal P, Said M, Molnár I, Doležel J (2023). Flow cytometric analysis and sorting of plant chromosomes. In: Heitkam T, Garcia S, eds. Plant Cytogenetics and Cytogenomics. New York: Humana. pp. 177-200. |
| [4] | Decaestecker W, Bollier N, Buono RA, Nowack MK, Jacobs TB (2022). Protoplast preparation and fluorescence- activated cell sorting for the evaluation of targeted mutagenesis in plant cells. In: Wang K, Zhang F, eds. Protoplast Technology. New York: Humana. pp. 205-221. |
| [5] | Doležel J, Číhalíková J, Lucretti S (1992). A high-yield procedure for isolation of metaphase chromosomes from root tips of Vicia faba L. Planta 188, 93-98. |
| [6] | Doležel J, Göhde W (1995). Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19, 103-106. |
| [7] | Doležel J, Greilhuber J, Suda J (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2, 2233-2244. |
| [8] | Doležel J, Lucretti S, Molnár I, Cápal P, Giorgi D (2021). Chromosome analysis and sorting. Cytometry A 99, 328- 342. |
| [9] | Dpooležel J, Binarová P, Lcretti S (1989). Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31, 113-120. |
| [10] | Fan W, Xia CJ, Wang SX, Liu J, Deng LJ, Sun SY, Wang XL (2022). Rhizobial infection of 4C cells triggers their endoreduplication during symbiotic nodule development in soybean. New Phytol 234, 1018-1030. |
| [11] | Galbraith DW, Afonso CL, Harkins KR (1984). Flow sorting and culture of protoplasts: conditions for high-frequency recovery, growth and morphogenesis from sorted protoplasts of suspension cultures of nicotiana. Plant Cell Rep 3, 151-155. |
| [12] | Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049-1051. |
| [13] | Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O'Shaughnessy AL, Lambert GM, Araúzo- Bravo MJ, Lee J, Fishman M, Robbins GE, Lin XY, Venepally P, Badger JH, Galbraith DW, Gage FH, Lasken RS (2013). RNA-sequencing from single nuclei. Proc Natl Acad Sci USA 110, 19802-19807. |
| [14] | Guedes JG, Guimaraes AL, Carqueijeiro I, Gardner R, Bispo C, Sottomayor M (2022). Isolation of specialized plant cells by fluorescence-activated cell sorting. In: Fett-Neto AG,ed. Plant Secondary Metabolism Engineering. New York: Humana. pp. 193-200. |
| [15] | Guillotin B, Rahni R, Passalacqua M, Mohammed MA, Xu XS, Raju SK, Ramírez CO, Jackson D, Groen SC, Gillis J, Birnbaum KD (2023). A pan-grass transcriptome reveals patterns of cellular divergence in crops. Nature 617, 785-791. |
| [16] | Hang HY, Liu CC, Ren DD (2019). Development, application and prospection of flow cytometry. China Biotechnol 39(9), 68-83. (in Chinese) |
| 杭海英, 刘春春, 任丹丹 (2019). 流式细胞术的发展、应用及前景. 中国生物工程杂志 39(9), 68-83. | |
| [17] | Kamentsky LA, Melamed MR, Derman H (1965). Spectrophotometer: new instrument for ultrarapid cell analysis. Science 150, 630-631. |
| [18] | Kawakatsu T, Stuart T, Valdes M, Breakfield N, Schmitz RJ, Nery JR, Urich MA, Han XW, Lister R, Benfey PN, Ecker JR (2016). Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants 2, 16058. |
| [19] | Li F, Hu Y, Wang F, Zhang Z, Liu XL, Bai SL, He YK (2010). Flow cytometry sorting of early developmental non- hair cells in roots of Arabidopsis thaliana. Chin Bull Bot 45, 460-465. (in Chinese) |
| 李斐, 胡勇, 王帆, 张珍, 刘祥林, 白素兰, 何奕昆 (2010). 利用流式细胞仪分选拟南芥根尖发育早期非根毛细胞. 植物学报 45, 460-465. | |
| [20] | Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HCW, Zhang YS, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16, 1295-1310. |
| [21] | Loureiro J, Rodriguez E, Doležel J, Santos C (2007). Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100, 875-888. |
| [22] | Marx V (2016). Plants: a tool box of cell-based assays. Nat Methods 13, 551-554. |
| [23] | Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23, 1249-1263. |
| [24] | Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, Iino T, Arai F, Endo T, Fujiwaki Y, Fukuzawa H, Hase M, Hayakawa T, Hiramatsu K, Hoshino Y, Inaba M, Ito T, Karakawa H, Kasai Y, Koizumi K, Lee S, Lei C, Li M, Maeno T, Matsusaka S, Murakami D, Nakagawa A, Oguchi Y, Oikawa M, Ota T, Shiba K, Shintaku H, Shirasaki Y, Suga K, Suzuki Y, Suzuki N, Tanaka Y, Tezuka H, Toyokawa C, Yalikun Y, Yamada M, Yamagishi M, Yamano T, Yasumoto A, Yatomi Y, Yazawa M, Di Carlo D, Hosokawa Y, Uemura S, Ozeki Y, Goda K (2018). Intelligent image-activated cell sorting. Cell 175, 266-276. |
| [25] | Ortiz-Ramírez C, Arevalo ED, Xu XS, Jackson DP, Birnbaum KD (2018). An efficient cell sorting protocol for maize protoplasts. Curr Protoc Plant Biol 3, e20072. |
| [26] | Otto FJ (1992). Preparation and staining of cells for high- resolution DNA analysis. In: Radbruch A, ed. Flow Cytometry and Cell Sorting. Berlin: Springer. pp. 65-68. |
| [27] | Petrovská B, Jeřábková H, Chamrád I, Vrána J, Lenobel R, Uřinovská J, Šebela M, Doležel J (2014). Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet Genome Res 143, 78-86. |
| [28] | Pfosser M, Heberle-Bors E, Amon A, Lelley T (1995). Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat- rye addition lines. Cytometry 21, 387-393. |
| [29] | Reichard A, Asosingh K (2019). Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry A 95, 219-226. |
| [30] | Šafář J, Bartoš J, Janda J, Bellec A, Kubaláková M, Valárik M, Pateyron S, Weiserová J, Tušková R, Číhalíková J, Vrána J, Šimková H, Faivre-Rampant P, Sourdille P, Caboche M, Bernard M, Doležel J, Chalhoub B (2004). Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J 39, 960-968. |
| [31] | Said M, Kubaláková M, Karafiátová M, Molnár I, Doležel J, Vrána J (2019). Dissecting the complex genome of crested wheatgrass by chromosome flow sorting. Plant Genome 12, 180096. |
| [32] | Schraivogel D, Kuhn TM, Rauscher B, Rodríguez-Martínez M, Paulsen M, Owsley K, Middlebrook A, Tischer C, Ramasz B, Ordoñez-Rueda D, Dees M, Cuylen- Haering S, Diebold E, Steinmetz LM (2022). High-speed fluorescence image-enabled cell sorting. Science 375, 315- 320. |
| [33] | Shapiro HM (2003). Practical Flow Cytometry, 4th edn. New York: Wiley-Liss. pp. 73-100. |
| [34] | Sliwinska E, Loureiro J, Leitch IJ, Šmarda P, Bainard J, Bureš P, Chumová Z, Horová L, Koutecký P, Lučanová M, Trávníček P, Galbraith DW (2022). Application-based guidelines for best practices in plant flow cytometry. Cytometry A 101, 749-781. |
| [35] | Smith JP, Sheffield NC (2020). Analytical approaches for ATAC-seq data analysis. Curr Protoc Hum Genet 106, e101. |
| [36] | Sun GL, Xia MZ, Li JP, Ma W, Li QZ, Xie JJ, Bai SL, Fang SS, Sun T, Feng XL, Guo GH, Niu YL, Hou JY, Ye WL, Ma JC, Guo SY, Wang HL, Long Y, Zhang XB, Zhang JL, Zhou H, Li BZ, Liu J, Zou CS, Wang H, Huang JL, Galbraith DW, Song CP (2022). The maize single-nucleus transcriptome comprehensively describes signaling networks governing movement and development of grass stomata. Plant Cell 34, 1890-1911. |
| [37] | Taher TEI (2017). Monitoring promoter activity by flow cytometry. In: Gould D, ed. Mammalian Synthetic Promoters. New York: Humana. pp. 65-73. |
| [38] | The International Wheat Genome Sequencing Consortium IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. |
| [39] | Xu XS, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu ZF, Wang LY, Fox N, Wang XF, Drenkow J, Luo AD, Char SN, Yang B, Sylvester AW, Gingeras TR, Schmitz RJ, Ware D, Lipka AE, Gillis J, Jackson D (2021). Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56, 557-568. |
| [40] | Zhang JD, Feng M (2023). A plant sample optimal pretreatment for flow cytometric analysis. Chin Bull Bot 58, 285-297. (in Chinese) |
| 张晋丹, 冯旻 (2023). 一种提升流式细胞术分析效果的前处理方法. 植物学报 58, 285-297. | |
| [41] | Zhu T, Liao KY, Zhou RF, Xia CJ, Xie WB (2020). ATAC- seq with unique molecular identifiers improves quantification and footprinting. Commun Biol 3, 675. |
| [42] | Zhu T, Xia CJ, Yu RR, Zhou XK, Xu XB, Wang L, Zong ZX, Yang JJ, Liu YM, Ming LC, You YX, Chen DJ, Xie WB (2024). Comprehensive mapping and modelling of the rice regulome landscape unveils the regulatory architecture underlying complex traits. Nat Commun 15, 6562. |
| [1] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [4] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [5] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [14] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [15] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||