

植物学报 ›› 2022, Vol. 57 ›› Issue (6): 826-836.DOI: 10.11983/CBB22159 cstr: 32102.14.CBB22159
所属专题: 饲草生物学专辑 (2023年58卷2期、2022年57卷6期); 大食物观; 粮食安全
曹丽雯1, 卢蕊2, 范吉标3, 胡龙兴2,*( ), 陈良1,*(
), 陈良1,*( )
)
收稿日期:2022-07-19
接受日期:2022-10-24
出版日期:2022-11-01
发布日期:2022-11-18
通讯作者:
胡龙兴,陈良
作者简介:chenliang888@wbgcas.cn基金资助:
Liwen Cao1, Rui Lu2, Jibiao Fan3, Longxing Hu2,*( ), Liang Chen1,*(
), Liang Chen1,*( )
)
Received:2022-07-19
Accepted:2022-10-24
Online:2022-11-01
Published:2022-11-18
Contact:
Longxing Hu,Liang Chen
摘要: 饲草是发展草食畜牧业的基石。然而, 现阶段由于饲草品种匮乏以及饲草种植结构单一引起的饲草短缺已成为饲草产业的一大瓶颈问题。因此, 在大力发展传统优质饲草的基础上, 亟须挖掘新型饲草的生产潜力。新型饲草是指在产量、营养品质、适应性和抗逆性等单方面或多方面较传统饲草具有明显优势、近年来饲用价值才被开发利用的饲草。该文以狗牙根(Cynodon dactylon)、小黑麦(×Triticosecale Wittmack)、藜麦(Chenopodium quinoa)、饲用油菜(Brassica napus)、籽粒苋、田菁(Sesbania cannabina)和野大豆(Glycine soja)等为主要对象, 系统梳理了新型饲草的国内外研究现状与发展趋势, 分析了我国在该研究领域的核心竞争力, 探讨了新型饲草育种中存在的重要基础生物学问题, 并提出了推动新型饲草产业健康发展的策略和建议, 以期促进新型饲草的种业创新和饲草产业的可持续发展, 保障国家大粮食安全。
曹丽雯, 卢蕊, 范吉标, 胡龙兴, 陈良. 新型饲草开发利用的基础生物学问题. 植物学报, 2022, 57(6): 826-836.
Liwen Cao, Rui Lu, Jibiao Fan, Longxing Hu, Liang Chen. The Fundamental Biological Problems in the Development and Utilization of New Forage Grass. Chinese Bulletin of Botany, 2022, 57(6): 826-836.

图1 常见新型饲草饲用狗牙根(A)、籽粒苋(B)、藜麦(C)、田菁(D)、饲用油菜(E)和小黑麦(F) Bars=10 cm
Figure 1 New forage grasses bermudagrass (A), grain amaranth (B), quinoa (C), sesban (D), forage rapeseed (E), and triticale (F) Bars=10 cm
| 饲草 | 鲜草产量 (kg?667m-2) | 粗蛋白 含量(%) | 粗纤维 含量(%) | 中性洗涤纤维含量(%) | 酸性洗涤纤维含量(%) | 粗脂肪 含量(%) | 年份 | 产地 | 参考文献 |
|---|---|---|---|---|---|---|---|---|---|
| 狗牙根 (岸杂1号) | 1675 (1茬) | 13.89 | 32.56 | \ | \ | 3.54 | 2013 | 广西 | 2017 |
| 籽粒苋 (茎叶) | 8667 | 16.00-23.00 | \ | \ | \ | 3.40 | \ | \ | 2019 |
| 小黑麦 (中饲828) | 2271-3715 | 15.60-18.10 | \ | 53.50-57.10 | 31.50-34.30 | 17.00-18.50 | 2019 | 天津 | 2021 |
| 饲用油菜(饲油1号) | 3586 | 23.46 | 11.65 | \ | \ | 4.03 | 2005-2006 | 甘肃 | 2021 |
| 藜麦 (中藜1号) | 3200 | 15.60-16.80 | \ | 34.50-35.20 | 45.20-46.10 | \ | 2014-2017 | 内蒙古 | 2022 |
| 野大豆 | 2700 (3茬) | 16.90-17.90 | 27.20-29.30 | \ | \ | \ | 2013 | 安徽 | 2014 |
| 田菁 | 2150-2400 | 12.20-16.70 | \ | \ | \ | \ | \ | \ | 1993 |
| 苜蓿 | 514 | 19 | \ | 35 | 45 | 28 | 2020 | 全国 平均 | 2022 |
| 青贮玉米 | 1050 | 8 | \ | 32 | 54 | 26 | 2020 | 全国 平均 | 2022 |
表1 不同新型饲草营养成分和产量统计
Table 1 The summary of forage nutritional composition and yield of new forage grasses
| 饲草 | 鲜草产量 (kg?667m-2) | 粗蛋白 含量(%) | 粗纤维 含量(%) | 中性洗涤纤维含量(%) | 酸性洗涤纤维含量(%) | 粗脂肪 含量(%) | 年份 | 产地 | 参考文献 |
|---|---|---|---|---|---|---|---|---|---|
| 狗牙根 (岸杂1号) | 1675 (1茬) | 13.89 | 32.56 | \ | \ | 3.54 | 2013 | 广西 | 2017 |
| 籽粒苋 (茎叶) | 8667 | 16.00-23.00 | \ | \ | \ | 3.40 | \ | \ | 2019 |
| 小黑麦 (中饲828) | 2271-3715 | 15.60-18.10 | \ | 53.50-57.10 | 31.50-34.30 | 17.00-18.50 | 2019 | 天津 | 2021 |
| 饲用油菜(饲油1号) | 3586 | 23.46 | 11.65 | \ | \ | 4.03 | 2005-2006 | 甘肃 | 2021 |
| 藜麦 (中藜1号) | 3200 | 15.60-16.80 | \ | 34.50-35.20 | 45.20-46.10 | \ | 2014-2017 | 内蒙古 | 2022 |
| 野大豆 | 2700 (3茬) | 16.90-17.90 | 27.20-29.30 | \ | \ | \ | 2013 | 安徽 | 2014 |
| 田菁 | 2150-2400 | 12.20-16.70 | \ | \ | \ | \ | \ | \ | 1993 |
| 苜蓿 | 514 | 19 | \ | 35 | 45 | 28 | 2020 | 全国 平均 | 2022 |
| 青贮玉米 | 1050 | 8 | \ | 32 | 54 | 26 | 2020 | 全国 平均 | 2022 |
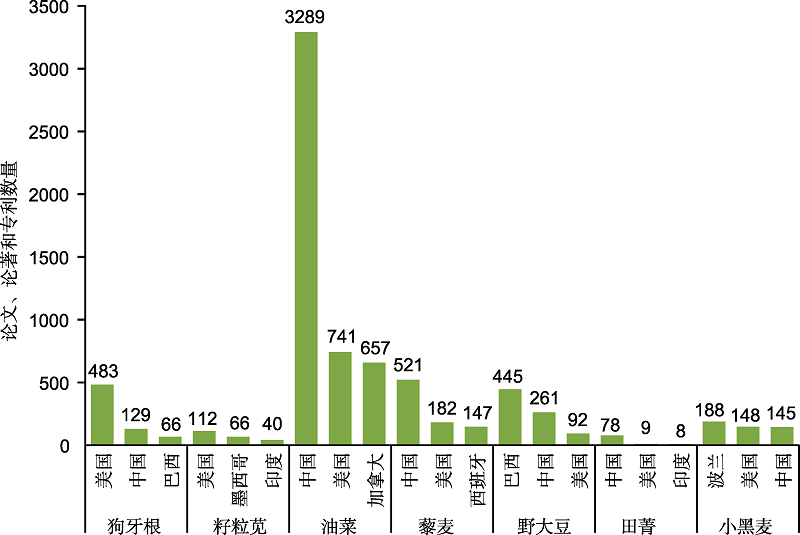
图2 近5年发表常见新型饲草相关文章、专利和论著数量排名前三的国家
Figure 2 Top three countries in the number of published papers, patents, and books about new forage grasses in the past five years
| 研究方向 | 研究单位 | 研究成果 |
|---|---|---|
| 狗牙根新品种培育 | 新疆农业大学 | 新农1号、新农2号、新农3号和喀什狗牙根 |
| 江苏省中国科学院植物研究所 | 南京狗牙根、阳江狗牙根、苏植2号和关中 | |
| 四川农业大学 | 川南和川西 | |
| 河北农业大学 | 保定狗牙根和邯郸狗牙根 | |
| 中国热带农业科学院热带作物品种资源研究所 | 桂南 | |
| 湖北省农业科学院畜牧兽医研究所 | 鄂引3号 | |
| 鲁东大学 | 鲁滨2号 | |
| 狗牙根复杂生物学性状调控机制研究、基因组破译、转化体系建立 | 中国科学院武汉植物园 | 挖掘鉴定了CdWRKY2、CdWRKY50和CdERF1等多个狗牙根抗寒、耐盐基因; 在狗牙根中建立了发根农杆菌介导的转化体系(Hu et al., |
| 华南农业大学 | 发掘鉴定了CdSMT1和CdtNF-YC1等多个狗牙根抗旱、耐盐基因(Chen et al., | |
| 南京农业大学 | 发掘鉴定了CtHsfA2b等耐热基因(Wang et al., | |
| 中国农业大学 | 完成非洲狗牙根基因组组装(Cui et al., | |
| 扬州大学 | 完成普通狗牙根基因组组装及全长转录组测序(Zhang et al., | |
| 鲁东大学 | 建立了根癌农杆菌介导的狗牙根遗传转化体系(Xu et al., | |
| 江苏省中国科学院植物研究所 | 在狗牙根中建立了病毒诱导的基因沉默体系(Zhang et al., | |
| 华中农业大学 | 测定狗牙根在非生物胁迫下的代谢组、转录组和蛋白质组并进行整合分析(Ye et al., | |
| 湖南农业大学 | 发现土壤养分影响狗牙根对镉的吸收、转运和耐受性(Chen et al., | |
| 四川农业大学 | 比较鉴定了狗牙根庇荫能力(Cao et al., |
表2 中国狗牙根研究团队核心竞争力分析
Table 2 Core competing capability of bermudagrass research groups in China
| 研究方向 | 研究单位 | 研究成果 |
|---|---|---|
| 狗牙根新品种培育 | 新疆农业大学 | 新农1号、新农2号、新农3号和喀什狗牙根 |
| 江苏省中国科学院植物研究所 | 南京狗牙根、阳江狗牙根、苏植2号和关中 | |
| 四川农业大学 | 川南和川西 | |
| 河北农业大学 | 保定狗牙根和邯郸狗牙根 | |
| 中国热带农业科学院热带作物品种资源研究所 | 桂南 | |
| 湖北省农业科学院畜牧兽医研究所 | 鄂引3号 | |
| 鲁东大学 | 鲁滨2号 | |
| 狗牙根复杂生物学性状调控机制研究、基因组破译、转化体系建立 | 中国科学院武汉植物园 | 挖掘鉴定了CdWRKY2、CdWRKY50和CdERF1等多个狗牙根抗寒、耐盐基因; 在狗牙根中建立了发根农杆菌介导的转化体系(Hu et al., |
| 华南农业大学 | 发掘鉴定了CdSMT1和CdtNF-YC1等多个狗牙根抗旱、耐盐基因(Chen et al., | |
| 南京农业大学 | 发掘鉴定了CtHsfA2b等耐热基因(Wang et al., | |
| 中国农业大学 | 完成非洲狗牙根基因组组装(Cui et al., | |
| 扬州大学 | 完成普通狗牙根基因组组装及全长转录组测序(Zhang et al., | |
| 鲁东大学 | 建立了根癌农杆菌介导的狗牙根遗传转化体系(Xu et al., | |
| 江苏省中国科学院植物研究所 | 在狗牙根中建立了病毒诱导的基因沉默体系(Zhang et al., | |
| 华中农业大学 | 测定狗牙根在非生物胁迫下的代谢组、转录组和蛋白质组并进行整合分析(Ye et al., | |
| 湖南农业大学 | 发现土壤养分影响狗牙根对镉的吸收、转运和耐受性(Chen et al., | |
| 四川农业大学 | 比较鉴定了狗牙根庇荫能力(Cao et al., |
| [1] | 陈其鲜, 毛万湖, 崔小茹, 孙多鑫 (2007). 饲草油菜新品种饲油1号的特征特性及栽培要点. 农业科技通讯 (8), 56. |
| [2] | 陈幼春, 初克森 (2019). 发展籽粒苋助力畜牧业可持续发展. 中国畜牧业 (22), 50-51. |
| [3] | 程皇座, 陈国福 (2019). 籽粒苋的饲用价值及其在猪生产中的应用. 养殖与饲料 (6), 60-62. |
| [4] | 崔力航, 苏惟真, 宗筱雯, 王亚坤 (2022). 2021年中国乳业贸易发展趋势与未来展望. 乳品与人类 (3), 4-17. |
| [5] | 李福岭 (1993). 田菁的饲料利用价值及饲喂试验. 饲料研究 (5), 30-31. |
| [6] | 刘瑞香, 郭占斌, 李进才, 任贵兴, 杨修仕, 秦培友 (2022). 中藜1号藜麦新品种选育及营养价值研究. 干旱区资源与环境 36(3), 166-170. |
| [7] | 刘伟, 张新全, 李芳, 马啸, 范彦 (2007). 西南区野生狗牙根遗传多样性的ISSR标记与地理来源分析. 草业学报 16(3), 55-61. |
| [8] | 孟春花, 张建丽, 钱勇, 王飞, 石祖梁, 仲跻峰 (2021). 油菜秸秆饲料化利用的研究进展. 江苏农业科学 49(16), 26-31. |
| [9] | 彭丽娟, 梁辛, 杨承剑, 韦升菊, 李丽莉, 罗华, 诸葛莹, 李舒露 (2017). 岸杂1号狗牙根牧草引种观察及其营养价值测定. 草学 (1), 45-48. |
| [10] | 孙国庆, 马健, 都文, 王雅晶, 曹志军, 李胜利, 余雄, 王宇, 雷小英, 马亚宾 (2017). 饲粮中添加籽粒苋对泌乳奶牛瘤胃发酵、血液指标和生产性能的影响. 动物营养学报 29, 1652-1660. |
| [11] | 王凤行 (2021). 两种饲用型小黑麦产量及其营养品质的研究. 中国农技推广 37(6), 38-39. |
| [12] | 王旭 (2022). 饲草产业发展迎来战略机遇期. 中国畜牧业 (8), 16-23. |
| [13] | 王艳荣, 王鸿升, 张海棠, 崔艳红 (2011). 优质饲用植物——籽粒苋的研究进展. 北方牧业 (3), 27. |
| [14] |
谢华玲, 杨艳萍, 董瑜, 王台 (2021). 苜蓿国际发展态势分析. 植物学报 56, 740-750.
DOI |
| [15] | 周芬, 陈胜, 李杨 (2014). 合肥市野生大豆的饲用价值研究. 安徽农业科学 42, 1066-1067, 1070. |
| [16] | Burton GW (2001). Tifton 85 bermudagrass-early history of its creation, selection, and evaluation. Crop Sci 41, 5-6. |
| [17] | Cao HS, Zhuo L, Su Y, Sun LX, Wang XM (2016). Non- specific phospholipase C1 affects silicon distribution and mechanical strength in stem nodes of rice. Plant J 86, 308-321. |
| [18] | Cao L, Yu Y, Ding XD, Zhu D, Yang F, Liu BD, Sun XL, Duan XB, Yin KD, Zhu YM (2017). The Glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol Biol 95, 253-268. |
| [19] | Cao YQ, Yang K, Liu W, Feng GY, Peng Y, Li Z (2022). Adaptive responses of common and hybrid bermudagrasses to shade stress associated with changes in morphology, photosynthesis, and secondary metabolites. Front Plant Sci 13, 817105. |
| [20] |
Castellanos-Arévalo AP, Estrada-Luna AA, Cabrera-Ponce JL, Valencia-Lozano E, Herrera-Ubaldo H, de Folter S, Blanco-Labra A, Délano-Frier JP (2020). Agrobacterium rhizogenes-mediated transformation of grain (Amaranthus hypochondriacus) and leafy (A. hybridus) amaranths. Plant Cell Rep 39, 1143-1160.
DOI PMID |
| [21] |
Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950-953.
DOI PMID |
| [22] |
Chen B, Tan SD, Zeng QR, Wang AD, Zheng HB (2019). Soil nutrient heterogeneity affects the accumulation and transfer of cadmium in bermuda grass (Cynodon dactylon (L.) pers.). Chemosphere 221, 342-348.
DOI PMID |
| [23] | Chen M, Chen JJ, Luo N, Qu RD, Guo ZF, Lu SY (2018). Cholesterol accumulation by suppression of SMT1 leads to dwarfism and improved drought tolerance in herbaceous plants. Plant Cell Environ 41, 1417-1426. |
| [24] |
Chen M, Zhao YJ, Zhuo CL, Lu SY, Guo ZF (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol J 13, 482-491.
DOI PMID |
| [25] | Cui FC, Taier G, Li ML, Dai XX, Hang N, Zhang XZ, Wang XF, Wang KH (2021). The genome of the warm-season turfgrass African bermudagrass (Cynodon transvaalensis). Hortic Res 8, 93. |
| [26] | Fang ZF, Liu CL, Ye TT, Huang C, Zhang ZW, Wang YP, Chan ZL (2022). Integrated transcriptome and proteome analyses provide insight into abiotic stress crosstalks in bermudagrass. Environ Exp Bot 199, 104864. |
| [27] | Feng X, Feng P, Yu HL, Yu XY, Sun Q, Liu SY, Minh TN, Chen J, Wang D, Zhang Q, Cao L, Zhou CM, Li Q, Xiao JL, Zhong SH, Wang AX, Wang LJ, Pan HY, Ding XD (2020). GsSnRK1 interplays with transcription factor GsERF7 from wild soybean to regulate soybean stress resistance. Plant Cell Environ 43, 1192-1211. |
| [28] | Hu ZR, Huang XB, Amombo E, Liu A, Fan JB, Bi AY, Ji K, Xin HP, Chen L, Fu JM (2020). The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci 294, 110432. |
| [29] | Huang XB, Amee M, Chen L (2021). Bermudagrass CdWRKY50 gene negatively regulates plants’ response to salt stress. Environ Exp Bot 188, 104513. |
| [30] | Huang XB, Cao LW, Fan JB, Ma GJ, Chen L (2022). CdWRKY2-mediated sucrose biosynthesis and CBF-signaling pathways coordinately contribute to cold tolerance in bermudagrass. Plant Biotechnol J 20, 660-675. |
| [31] | Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45, 462-465. |
| [32] | Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, Kharbatia NM, Rupper RR, Sharp AR, Dally N, Boughton BA, Woo YH, Gao G, Schijlen EGWM, Guo XJ, Momin AA, Negrão S, Al-Babili S, Gehring C, Roessner U, Jung C, Murphy K, Arold ST, Gojobori T, Van Der Linden CG, Van Loo EN, Jellen EN, Maughan PJ, Tester M (2017). The genome of Chenopodium quinoa. Nature 542, 307-312. |
| [33] | Ji H, Kim SR, Kim YH, Kim H, Eun MY, Jin ID, Cha YS, Yun DW, Ahn BO, Lee MC, Lee GS, Yoon UH, Lee JS, Lee YH, Suh SC, Jiang WZ, Yang JI, Jin P, McCouch SR, An G, Koh HJ (2010). Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61, 96-106. |
| [34] | Jiang LY, Ma X, Zhao SS, Tang YY, Liu FX, Gu P, Fu YC, Zhu ZF, Cai HW, Sun CQ, Tan LB (2019). The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell 31, 17-36. |
| [35] | Jin T, Sun YY, Shan Z, He JB, Wang N, Gai JY, Li Y (2021). Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol J 19, 1155-1169. |
| [36] |
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392-1396.
DOI PMID |
| [37] |
Lee J, Park JJ, Kim SL, Yim J, An G (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65, 487-499.
DOI PMID |
| [38] |
Li C, Zhou A, Sang T (2006). Rice domestication by reducing shattering. Science 311, 1936-1939.
DOI PMID |
| [39] |
Li GW, Wang LJ, Yang JP, He H, Jin HB, Li XM, Ren TH, Ren ZL, Li F, Han X, Zhao XG, Dong LL, Li YW, Song ZP, Yan ZH, Zheng NN, Shi CL, Wang ZH, Yang SL, Xiong ZJ, Zhang ML, Sun GH, Zheng X, Gou MY, Ji CM, Du JK, Zheng HK, Doležel J, Deng XW, Stein N, Yang QH, Zhang KP, Wang DW (2021). A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat Genet 53, 574-584.
DOI PMID |
| [40] | Li HG, Cheng X, Zhang LP, Hu JH, Zhang FG, Chen BY, Xu K, Gao GZ, Li H, Li LX, Huang Q, Li ZY, Yan GX, Wu XM (2018). An integration of genome-wide association study and gene co-expression network analysis identifies candidate genes of stem lodging-related traits in Brassica napus. Front Plant Sci 9, 796. |
| [41] |
Lightfoot DJ, Jarvis DE, Ramaraj T, Lee R, Jellen EN, Maughan PJ (2017). Single-molecule sequencing and Hi- C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol 15, 74.
DOI PMID |
| [42] | Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766-770. |
| [43] |
Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843-853.
DOI PMID |
| [44] | Lin ZW, Griffith ME, Li XR, Zhu ZF, Tan LB, Fu YC, Zhang WX, Wang XK, Xie DX, Sun CQ (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226, 11-20. |
| [45] | Lin ZW, Li XR, Shannon LM, Yeh CT, Wang ML, Bai GH, Peng Z, Li JR, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu JM (2012). Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44, 720-724. |
| [46] | Liu MY, Sun TX, Liu CL, Zhang H, Wang WL, Wang YP, Xiang L, Chan ZL (2022). Integrated physiological and transcriptomic analyses of two warm- and cool-season turfgrass species in response to heat stress. Plant Physiol Biochem 170, 275-286. |
| [47] |
Lv SW, Wu WG, Wang MH, Meyer RS, Ndjiondjop MN, Tan LB, Zhou HY, Zhang JW, Fu YC, Cai HW, Sun CQ, Wing RA, Zhu ZF (2018). Genetic control of seed shattering during African rice domestication. Nat Plants 4, 331-337.
DOI PMID |
| [48] | Ma X, Vaistij FE, Li Y, Jansen van Rensburg WS, Harvey S, Bairu MW, Venter SL, Mavengahama S, Ning ZM, Graham IA, Van Deynze A, Van De Peer Y, Denby KJ (2021). A chromosome-level Amaranthus cruentus genome assembly highlights gene family evolution and biosynthetic gene clusters that may underpin the nutritional value of this traditional crop. Plant J 107, 613-628. |
| [49] |
Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, Lerchl J, Landes A, Gatzmann F, Janssen A (2020). Draft genomes of Amaranthus tuberculatus, Amaranthus hybridus, and Amaranthus palmeri, Genome Biol Evol 12, 1988-1993.
DOI PMID |
| [50] | Munusamy U, Abdullah SNA, Aziz MA, Khazaai H (2013). Female reproductive system of Amaranthus as the target for Agrobacterium-mediated transformation. Adv Biosci Biotechnol 4, 28413. |
| [51] | Ning WF, Zhai H, Yu JQ, Liang S, Yang X, Xing XY, Huo JL, Pang T, Yang YL, Bai X (2017). Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol Breeding 37, 19. |
| [52] |
Rajani S, Sundaresan V (2001). The Arabidopsis myc/ bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11, 1914-1922.
DOI PMID |
| [53] | Robles P, Pelaz S (2004). Flower and fruit development in Arabidopsis thaliana. Int J Dev Biol 49, 633-643. |
| [54] | Shen XJ, Wang YY, Zhang YX, Guo W, Jiao YQ, Zhou XA (2018). Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int J Mol Sci 19, 3958. |
| [55] | Song JM, Guan ZL, Hu JL, Guo CC, Yang ZQ, Wang S, Liu DX, Wang B, Lu SP, Zhou R, Xie WZ, Cheng YF, Zhang YT, Liu KD, Yang QY, Chen LL, Guo L (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6, 34-45. |
| [56] | Sun PY, Zhang WH, Wang YH, He Q, Shu F, Liu H, Wang JM, Wang J, Yuan LP, Deng HF (2016). OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol 58, 836-847. |
| [57] | Wang XY, Huang WL, Yang ZM, Liu J, Huang BR (2016). Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci Rep 6, 28021. |
| [58] | Xu X, Liu WW, Liu XY, Cao YP, Li XN, Wang GY, Fu CX, Fu JM (2022). Genetic manipulation of bermudagrass photosynthetic biosynthesis using Agrobacterium-mediated transformation. Physiol Plant 174, e13710. |
| [59] | Ye TT, Wang YP, Feng YQ, Chan ZL (2021). Physiological and metabolomic responses of bermudagrass (Cynodon dactylon) to alkali stress. Physiol Plant 171, 22-33. |
| [60] | Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G (2014). The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J 79, 717-728. |
| [61] | Yu Y, Duan XB, Ding XD, Chen C, Zhu D, Yin KD, Cao L, Song XW, Zhu PH, Li Q, Nisa Z, Yu JY, Du JY, Song Y, Li HQ, Liu BD, Zhu YM (2017). A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol Biol 94, 509-530. |
| [62] | Zhang B, Chen S, Liu JX, Yan YB, Chen JB, Li DD, Liu JY (2022). A high-quality haplotype-resolved genome of common bermudagrass (Cynodon dactylon L.) provides insights into polyploid genome stability and prostrate growth. Front Plant Sci 13, 890980. |
| [63] | Zhang B, Liu JX, Wang XS, Wei ZW (2018a). Full-length RNA sequencing reveals unique transcriptome composition in bermudagrass. Plant Physiol Biochem 132, 95-103. |
| [64] | Zhang B, Shi JA, Chen JB, Li DD, Li JJ, Guo HL, Zong JQ, Wang Y, Guo AG, Liu JX (2016). Efficient virus-induced gene silencing in Cynodon dactylon and Zoysia japonica using rice tungro bacilliform virus vectors. Sci Hortic 207, 97-103. |
| [65] | Zhang Y, Xu AX, Lang LN, Wang Y, Liu X, Liang FH, Zhang BB, Qin MF, Dalelhan J, Huang Z (2018b). Genetic mapping of a lobed-leaf gene associated with salt tolerance in Brassica napus L. Plant Sci 269, 75-84. |
| [66] |
Zhong H, Guo QQ, Chen L, Ren F, Wang QQ, Zheng Y, Li XB (2012). Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep 31, 1991-2003.
DOI PMID |
| [67] | Zhou Y, Lu DF, Li CY, Luo JH, Zhu BF, Zhu JJ, Shangguan YY, Wang ZX, Sang T, Zhou B, Han B (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24, 1034-1048. |
| [68] | Zhuang YB, Wang XT, Li XC, Hu JM, Fan LC, Landis JB, Cannon SB, Grimwood J, Schmutz J, Jackson SA, Doyle JJ, Zhang XS, Zhang DJ, Ma JX (2022). Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat Plants 8, 233-244. |
| [1] | 邓娴, 李彤, 曹晓风. 基因编辑在饲草育种中的应用与展望[J]. 植物学报, 2023, 58(2): 233-240. |
| [2] | 王甜甜, 曹丽雯, 刘智全, 杨庆山, 陈良, 陈敏, 景海春. 黄河三角洲滨海草带建设的饲草基础生物学问题[J]. 植物学报, 2022, 57(6): 837-847. |
| [3] | 赵富伟, 蔡蕾, 臧春鑫. 遗传资源获取与惠益分享相关国际制度新进展[J]. 生物多样性, 2017, 25(11): 1147-1155. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||