

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (3): 343-349.DOI: 10.11983/CBB18106 cstr: 32102.14.CBB18106
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Xia Zhang1,Xiang Jing1,Guangcai Zhou2,Ying Bao1,*( )
)
Received:2018-04-26
Accepted:2018-12-10
Online:2019-05-01
Published:2019-11-24
Contact:
Ying Bao
Xia Zhang,Xiang Jing,Guangcai Zhou,Ying Bao. Phylogeny and Tissue-specific Expression of the GBSS Genes in Oryza officinalis[J]. Chinese Bulletin of Botany, 2019, 54(3): 343-349.
| Gene | Primer name | Primer sequence (5'-3') |
|---|---|---|
| GBSSI | GBSSI-F | AACGTGGCTGCTCCTTGAA |
| GBSSI-R | TTGGCAATAAGCCACACACA | |
| GBSSII | GBSSII-F | AGGCATCGAGGGTGAGGAG |
| GBSSII-R | CCATCTGGCCCACATCTCTA |
Table 1 Primer sequences of GBSSI and GBSSII
| Gene | Primer name | Primer sequence (5'-3') |
|---|---|---|
| GBSSI | GBSSI-F | AACGTGGCTGCTCCTTGAA |
| GBSSI-R | TTGGCAATAAGCCACACACA | |
| GBSSII | GBSSII-F | AGGCATCGAGGGTGAGGAG |
| GBSSII-R | CCATCTGGCCCACATCTCTA |
| Plants | Gene (GenBank/UniProt No.) | Sequence identity (%) | |
|---|---|---|---|
| Oryza officinalis | |||
| GBSSI | GBSSII | ||
| Oryza sativa subsp. japonica | GBSSI (XP_025882300.1) | 95.22 | 60.35 |
| GBSSII (XP_015647210.1) | 62.30 | 96.72 | |
| O. sativa subsp. indica | GBSSI (AAN77100.1) | 97.87 | 62.30 |
| GBSSII (ACY56079.1) | 62.46 | 97.04 | |
| Leersia perrieri | GBSSI (A0A0D9WLF6) * | 93.01 | 62.15 |
| GBSSII (A0A0D9WED4) * | 58.44 | 85.62 | |
| Zizania latifolia | GBSSI (ASSH01051725.1) ** | 90.80 | 62.01 |
| GBSSII (ASSH01023543.1) ** | 59.46 | 87.47 | |
| Brachypodium distachyon | GBSSI (XP_003557139.1) | 79.31 | 61.16 |
| GBSSII (XP_003569238.1) | 60.33 | 85.43 | |
| Hordeum vulgare | GBSSI (BAC41202.1) | 81.97 | 60.76 |
| GBSSII (BAJ99426.1) | 61.85 | 86.12 | |
| Setaria italica | GBSSI (AGW27658.1) | 86.72 | 61.90 |
| GBSSII (XP_004956034.1) | 62.40 | 87.34 | |
| Sorghum bicolor | GBSSI (XP_002436418.1) | 82.52 | 61.90 |
| GBSSII (XP_002461889.1) | 62.62 | 86.18 | |
| Triticum aestivum | GBSSI (XP_020146905.1) | 81.37 | 60.36 |
| GBSSII (AAG27624.1) | 61.79 | 86.09 | |
| Zea mays | GBSSI (NP_001105001) | 82.08 | 62.13 |
| GBSSII (NP_001334833) | 62.36 | 85.88 | |
| Musa acuminata | GBSS1 (KF512020.1) | 66.39 | 65.35 |
| GBSS2 (KF512021.1) | 67.96 | 63.99 | |
| GBSS3 (KF512023.1) | 63.93 | 62.91 | |
| Arabidopsis thaliana | GBSS (XP_020866584.1) | 62.48 | 63.88 |
| Gossypium raimondii | GBSS1 (XP_012474755.1) | 63.89 | 66.01 |
| GBSS2 (XP_012439861.1) | 63.11 | 66.77 | |
| GBSS3 (XP_012486622.1) | 62.75 | 64.01 | |
| Solanum lycopersicum | GBSS (NP_001311457.1) | 67.05 | 62.03 |
| Carica papaya | GBSS (XP_021900468.1) | 65.91 | 66.45 |
| S. tuberosum | GBSS (XP_006343763.1) | 62.89 | 67.27 |
| Vitis vinifera | GBSS1 (XP_010660257.1) | 65.52 | 66.23 |
| GBSS2 (XP_019081062.1) | 64.07 | 67.76 | |
| Amborella trichopoda | GBSS (XP_006837847.1) | 62.81 | 66.17 |
| Chlamydomonas reinhardtii | GBSS (XP_001697117.1) | 47.79 | 47.05 |
Table 2 Amino acid sequence identities of GBSSs between Oryza officinalis and other plants
| Plants | Gene (GenBank/UniProt No.) | Sequence identity (%) | |
|---|---|---|---|
| Oryza officinalis | |||
| GBSSI | GBSSII | ||
| Oryza sativa subsp. japonica | GBSSI (XP_025882300.1) | 95.22 | 60.35 |
| GBSSII (XP_015647210.1) | 62.30 | 96.72 | |
| O. sativa subsp. indica | GBSSI (AAN77100.1) | 97.87 | 62.30 |
| GBSSII (ACY56079.1) | 62.46 | 97.04 | |
| Leersia perrieri | GBSSI (A0A0D9WLF6) * | 93.01 | 62.15 |
| GBSSII (A0A0D9WED4) * | 58.44 | 85.62 | |
| Zizania latifolia | GBSSI (ASSH01051725.1) ** | 90.80 | 62.01 |
| GBSSII (ASSH01023543.1) ** | 59.46 | 87.47 | |
| Brachypodium distachyon | GBSSI (XP_003557139.1) | 79.31 | 61.16 |
| GBSSII (XP_003569238.1) | 60.33 | 85.43 | |
| Hordeum vulgare | GBSSI (BAC41202.1) | 81.97 | 60.76 |
| GBSSII (BAJ99426.1) | 61.85 | 86.12 | |
| Setaria italica | GBSSI (AGW27658.1) | 86.72 | 61.90 |
| GBSSII (XP_004956034.1) | 62.40 | 87.34 | |
| Sorghum bicolor | GBSSI (XP_002436418.1) | 82.52 | 61.90 |
| GBSSII (XP_002461889.1) | 62.62 | 86.18 | |
| Triticum aestivum | GBSSI (XP_020146905.1) | 81.37 | 60.36 |
| GBSSII (AAG27624.1) | 61.79 | 86.09 | |
| Zea mays | GBSSI (NP_001105001) | 82.08 | 62.13 |
| GBSSII (NP_001334833) | 62.36 | 85.88 | |
| Musa acuminata | GBSS1 (KF512020.1) | 66.39 | 65.35 |
| GBSS2 (KF512021.1) | 67.96 | 63.99 | |
| GBSS3 (KF512023.1) | 63.93 | 62.91 | |
| Arabidopsis thaliana | GBSS (XP_020866584.1) | 62.48 | 63.88 |
| Gossypium raimondii | GBSS1 (XP_012474755.1) | 63.89 | 66.01 |
| GBSS2 (XP_012439861.1) | 63.11 | 66.77 | |
| GBSS3 (XP_012486622.1) | 62.75 | 64.01 | |
| Solanum lycopersicum | GBSS (NP_001311457.1) | 67.05 | 62.03 |
| Carica papaya | GBSS (XP_021900468.1) | 65.91 | 66.45 |
| S. tuberosum | GBSS (XP_006343763.1) | 62.89 | 67.27 |
| Vitis vinifera | GBSS1 (XP_010660257.1) | 65.52 | 66.23 |
| GBSS2 (XP_019081062.1) | 64.07 | 67.76 | |
| Amborella trichopoda | GBSS (XP_006837847.1) | 62.81 | 66.17 |
| Chlamydomonas reinhardtii | GBSS (XP_001697117.1) | 47.79 | 47.05 |
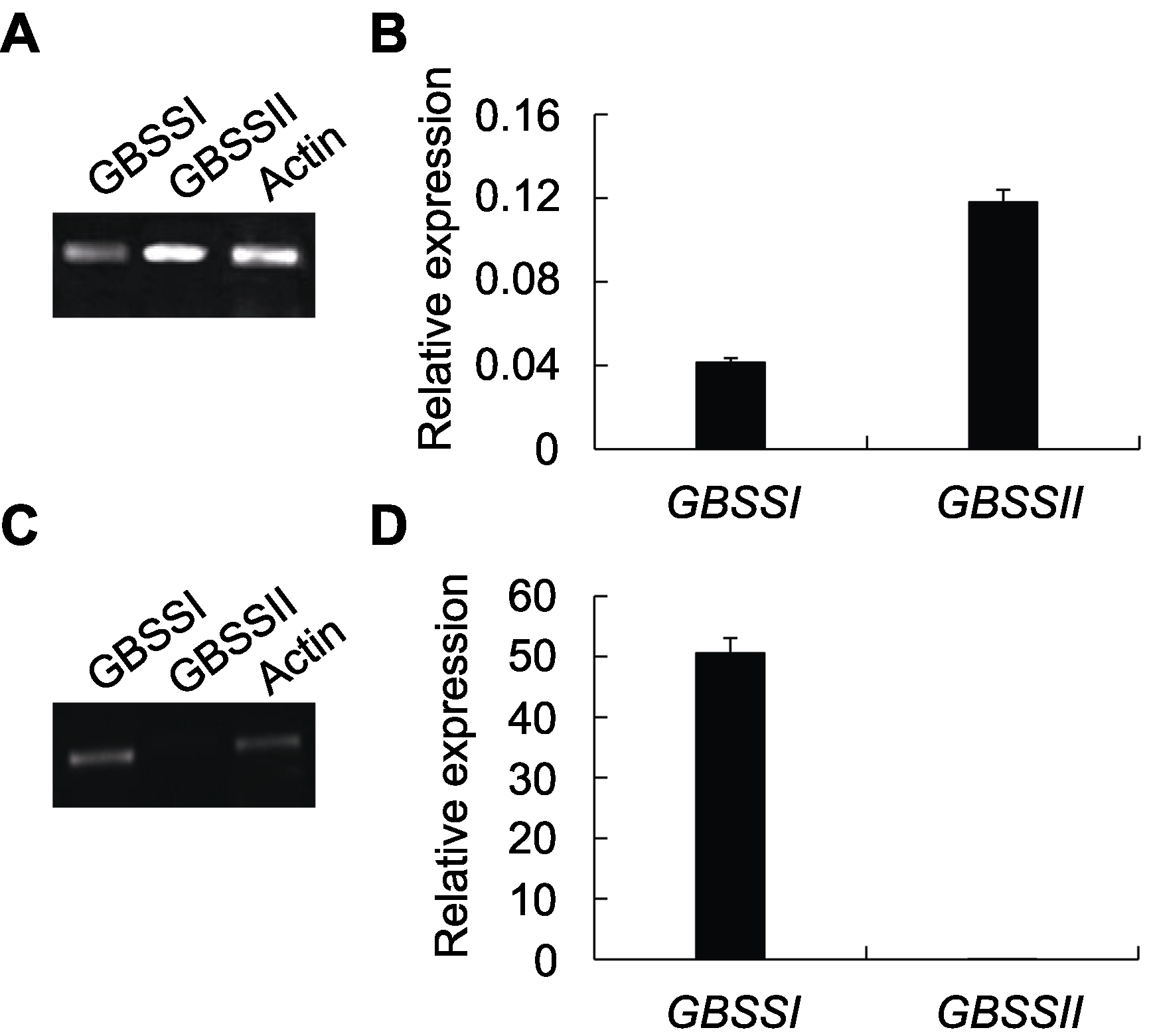
Figure 3 Relative expression of GBSSI and GBSSII in leaves and seeds of Oryza officinalis(A), (B) Amplification results of RT-PCR and qRT-PCR for GBSS genes in leaves; (C), (D) Amplification results of RT-PCR and qRT-PCR for GBSS genes in seeds.
| [1] |
包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报 50, 683-690.
DOI URL |
| [2] |
陈凤花, 王琳, 胡丽华 (2005). 实时荧光定量RT-PCR内参基因的选择. 临床检验杂志 23, 393-395.
DOI URL |
| [3] |
顾燕娟 (2006). 支链淀粉合成相关基因等位基因间的差异对稻米淀粉理化特性的影响. 硕士论文. 扬州: 扬州大学. pp. 3-8.
DOI URL |
| [4] |
王倩, 孙文静, 包颖 (2017). 植物颗粒结合淀粉合酶GBSS基因家族的进化. 植物学报 52, 179-187.
DOI URL |
| [5] |
杨学明 (2003). 几个重复序列在不同稻种中的分布及其与稻种分化关系的研究. 硕士论文. 扬州: 扬州大学. pp. 6-10.
DOI URL |
| [6] |
张鹏 (2008). 抑制淀粉分支酶类基因表达对稻米品质影响的研究. 硕士论文. 扬州: 扬州大学. pp. 3-8.
DOI URL |
| [7] | Bao Y, Xu S, Jing X, Meng L, Qin ZY (2015). De novo assembly and characterization ofOryza officinalis leaf transcriptome by using RNA-seq. Biomed Res Int 2015, 982065. |
| [8] |
Cheng J, Khan MA, Qiu WM, Li J, Zhou H, Zhang Q, Guo WW, Zhu TT, Peng JH, Sun FJ, Li SH, Korban SS, Han YP (2012). Diversification of genes encoding granulebound starch synthase in monocots and dicots is marked by multiple genome-wide duplication events.PLoS One 7, e30088.
DOI URL PMID |
| [9] |
Dian WM, Jiang HW, Wu P (2005). Evolution and expression analysis of starch synthase III and IV in rice.J Exp Bot 56, 623-632.
DOI URL PMID |
| [10] |
Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224.
DOI URL PMID |
| [11] |
Guindon S, Gascuel O (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Syst Biol 52, 696-704.
DOI URL PMID |
| [12] |
Ohdan T, Francisco Jr PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244.
DOI URL PMID |
| [13] | Shure M, Wessler S, Fedoroff N (1983). Molecular identification and isolation of thewaxy locus in maize. Cell 35, 225-233. |
| [14] | Van Harsselaar JK, Lorenz J, Senning M, Sonnewald U, Sonnewald S (2017). Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genomics 18, 37. |
| [15] |
Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264.
DOI URL |
| [16] | Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM (1990). Nucleotide sequence of ricewaxy gene. Nucleic Acids Res 18, 5898. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||