

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 350-359.DOI: 10.11983/CBB18205 cstr: 32102.14.CBB18205
胡建霖1,2,刘志芳1,2,慈秀芹1,李捷1,*
收稿日期:2018-09-27
接受日期:2019-02-11
出版日期:2019-05-01
发布日期:2019-11-24
通讯作者:
李捷
基金资助:Jianlin Hu1,2,Zhifang Liu1,2,Xiuqin Ci1,Jie Li1,*
Received:2018-09-27
Accepted:2019-02-11
Online:2019-05-01
Published:2019-11-24
Contact:
Jie Li
摘要: 龙脑香科植物是东南亚地区重要的热带木材来源树种, 对其开展DNA条形码评估在林业监管及森林资源保护等方面具有非常重要的实际应用价值。通过对龙脑香科植物样品进行rbcL、matK、trnL-trnF和ITS2四个片段的扩增和测序, 结合GenBank下载的数据, 共获得龙脑香科树种14属244种共计899条序列。通过比较4个片段的通用性、序列特征、种内和种间的遗传变异, 基于Best Match (BM)、Best Close Match (BCM)、相似性搜索算法(BLAST)和邻接树(NJ) 4种方法评估DNA条形码对于龙脑香科树种的鉴定能力。结果表明, ITS2在龙脑香科树种中鉴定效率最高, 通过优化的扩增体系能够从该科植物叶片中获得较高质量的ITS2片段; 叶绿体matK片段扩增和测序效率为100%, 且种内及种间遗传变异明显, 鉴定成功率高于其它叶绿体片段, 并据此提出ITS2和matK适合作为龙脑香科树种的DNA条形码。
胡建霖,刘志芳,慈秀芹,李捷. DNA条形码在热带龙脑香科树种鉴定中的应用. 植物学报, 2019, 54(3): 350-359.
Jianlin Hu,Zhifang Liu,Xiuqin Ci,Jie Li. Use of DNA Barcoding in Identifying Tropical Trees from Dipterocarpaceae. Chinese Bulletin of Botany, 2019, 54(3): 350-359.
| No. | Species | Voucher numbers | GenBank accession number | |||
|---|---|---|---|---|---|---|
| rbcL | matK | trnL-trnF | ITS2 | |||
| 1 | Dipterocarpus turbinatus C. F. Gaertner | JL5901 | MK030561 | MK051080 | MK051054 | MK051069 |
| 2 | D. turbinatus C. F. Gaertner | JL5902 | MK030562 | MK051081 | MK051055 | MK051070 |
| 3 | D. retusus Blume | JL9601 | MK030559 | MK051078 | MK051052 | MK193032 |
| 4 | D. retusus Blume | JL9602 | MK030560 | MK051079 | MK051053 | MK193031 |
| 5 | Hopea chinensis (Merr.) Hand.-Mazz. | JL110A | MK030563 | MK051082 | MK193024 | MK193026 |
| 6 | H. chinensis (Merr.) Hand.-Mazz. | JL110B | MK030564 | MK051083 | MK051059 | MK193025 |
| 7 | H. hainanensis Merrill & Chun | JL2201 | MK030565 | MK051084 | MK051060 | MK193029 |
| 8 | H. hainanensis Merrill & Chun | JL2202 | MK030566 | MK051085 | MK051061 | MK193028 |
| 9 | H. hainanensis Merrill & Chun | JL2203 | MK030567 | MK051086 | MK051062 | MK193030 |
| 10 | Parashorea chinensis H. Wang | JL6101 | MK030568 | MK051087 | MK051056 | MK051071 |
| 11 | P. chinensis H. Wang | JL6102 | MK030569 | MK051088 | MK051057 | MK051072 |
| 12 | Shorea assamica Dyer | JL3901 | MK030571 | MK051090 | MK051063 | - |
| 13 | S. assamica Dyer | JL3902 | MK030570 | MK051089 | MK051058 | MK051073 |
| 14 | Vatica diospyroides Symington | JL7601 | MK030572 | MK051091 | MK051064 | MK051074 |
| 15 | V. diospyroides Symington | JL7602 | MK030573 | MK051092 | MK193023 | MK051075 |
| 16 | V. guangxiensis X. L. Mo | JL4801 | MK030574 | MK051093 | MK051065 | MK051076 |
| 17 | V. guangxiensis X. L. Mo | JL4802 | MK030575 | MK051094 | MK051066 | MK051077 |
| 18 | V. mangachapoi Blanco | JL111A | MK030576 | MK051095 | MK051067 | - |
| 19 | V. mangachapoi Blanco | JL111B | MK030577 | MK051096 | MK051068 | MK193027 |
表1 实验材料
Table 1 Information of experimental materials
| No. | Species | Voucher numbers | GenBank accession number | |||
|---|---|---|---|---|---|---|
| rbcL | matK | trnL-trnF | ITS2 | |||
| 1 | Dipterocarpus turbinatus C. F. Gaertner | JL5901 | MK030561 | MK051080 | MK051054 | MK051069 |
| 2 | D. turbinatus C. F. Gaertner | JL5902 | MK030562 | MK051081 | MK051055 | MK051070 |
| 3 | D. retusus Blume | JL9601 | MK030559 | MK051078 | MK051052 | MK193032 |
| 4 | D. retusus Blume | JL9602 | MK030560 | MK051079 | MK051053 | MK193031 |
| 5 | Hopea chinensis (Merr.) Hand.-Mazz. | JL110A | MK030563 | MK051082 | MK193024 | MK193026 |
| 6 | H. chinensis (Merr.) Hand.-Mazz. | JL110B | MK030564 | MK051083 | MK051059 | MK193025 |
| 7 | H. hainanensis Merrill & Chun | JL2201 | MK030565 | MK051084 | MK051060 | MK193029 |
| 8 | H. hainanensis Merrill & Chun | JL2202 | MK030566 | MK051085 | MK051061 | MK193028 |
| 9 | H. hainanensis Merrill & Chun | JL2203 | MK030567 | MK051086 | MK051062 | MK193030 |
| 10 | Parashorea chinensis H. Wang | JL6101 | MK030568 | MK051087 | MK051056 | MK051071 |
| 11 | P. chinensis H. Wang | JL6102 | MK030569 | MK051088 | MK051057 | MK051072 |
| 12 | Shorea assamica Dyer | JL3901 | MK030571 | MK051090 | MK051063 | - |
| 13 | S. assamica Dyer | JL3902 | MK030570 | MK051089 | MK051058 | MK051073 |
| 14 | Vatica diospyroides Symington | JL7601 | MK030572 | MK051091 | MK051064 | MK051074 |
| 15 | V. diospyroides Symington | JL7602 | MK030573 | MK051092 | MK193023 | MK051075 |
| 16 | V. guangxiensis X. L. Mo | JL4801 | MK030574 | MK051093 | MK051065 | MK051076 |
| 17 | V. guangxiensis X. L. Mo | JL4802 | MK030575 | MK051094 | MK051066 | MK051077 |
| 18 | V. mangachapoi Blanco | JL111A | MK030576 | MK051095 | MK051067 | - |
| 19 | V. mangachapoi Blanco | JL111B | MK030577 | MK051096 | MK051068 | MK193027 |
| Barcodes | Primers sequence (5'-3') | Amplification protocol | References |
|---|---|---|---|
| rbcL | 1F: ATGTCACCACAAACAGAGACTAAAGC 724R: TCGCATGTACCTGCAGTAGC | 94°C 4 min; 94°C 30 s, 55°C 45 s, 72°C 1 min, 5 cycles; 94°C 30 s, 54°C 45 s, 72°C 10 min, 30 cycles; 72°C 10 min | Fay et al., 1997 Olmstead et al., 1992 |
| matK | 3F: CGTACAGTACTTTTGTGTTTACGA 1R: ACCCAGTCCATCTGGAAATCTTGG | 94°C 4 min; 94°C 30 s, 51°C 50 s, 72°C 50 s, 35 cycles; 72°C 10 min | Unpublished |
| 390F: CGATCTATTCATTCAATATTTC 1326R: TCTAGCACACGAAAGAAGT | 94°C 4 min; 94°C 30 s, 50°C 50 s, 72°C 50 s, 35 cycles; 72°C 10 min | Cuénoud et al., 2002 | |
| trnL-trnF | F: GGTTCAAGTCCCTCTATCCC R: ATTTGAACTGGTGACACGAG | 94°C 4 min; 94°C 30 s, 55°C 1 min, 72°C 1 min, 35 cycles; 72°C 5 min | Taberlet et al.,1991 |
| ITS2 | ITS2 2F: ATGCGATACTTGGTGTGAAT ITS2 3R: GACGCTTCTCCAGACTACAAT | 94°C 4 min; 94°C 30 s, 55°C 45 s, 72°C 45 s, 35 cycles; 72°C 10 min | Chen et al., 2010 Chen et al., 2010 |
| ITS3: GCATCGATGAAGAACGCAGC 26SE: TAGAATTCCCCGGTTCGCTCGCCGTTAC | 94°C 4 min; 94°C 30 s, 57°C 45 s, 72°C 45 s, 35 cycles; 72°C 10 min | White et al., 1990; Sun et al., 1994 |
表2 PCR扩增引物及程序
Table 2 Primer pairs used for PCR amplification and amplification protocol
| Barcodes | Primers sequence (5'-3') | Amplification protocol | References |
|---|---|---|---|
| rbcL | 1F: ATGTCACCACAAACAGAGACTAAAGC 724R: TCGCATGTACCTGCAGTAGC | 94°C 4 min; 94°C 30 s, 55°C 45 s, 72°C 1 min, 5 cycles; 94°C 30 s, 54°C 45 s, 72°C 10 min, 30 cycles; 72°C 10 min | Fay et al., 1997 Olmstead et al., 1992 |
| matK | 3F: CGTACAGTACTTTTGTGTTTACGA 1R: ACCCAGTCCATCTGGAAATCTTGG | 94°C 4 min; 94°C 30 s, 51°C 50 s, 72°C 50 s, 35 cycles; 72°C 10 min | Unpublished |
| 390F: CGATCTATTCATTCAATATTTC 1326R: TCTAGCACACGAAAGAAGT | 94°C 4 min; 94°C 30 s, 50°C 50 s, 72°C 50 s, 35 cycles; 72°C 10 min | Cuénoud et al., 2002 | |
| trnL-trnF | F: GGTTCAAGTCCCTCTATCCC R: ATTTGAACTGGTGACACGAG | 94°C 4 min; 94°C 30 s, 55°C 1 min, 72°C 1 min, 35 cycles; 72°C 5 min | Taberlet et al.,1991 |
| ITS2 | ITS2 2F: ATGCGATACTTGGTGTGAAT ITS2 3R: GACGCTTCTCCAGACTACAAT | 94°C 4 min; 94°C 30 s, 55°C 45 s, 72°C 45 s, 35 cycles; 72°C 10 min | Chen et al., 2010 Chen et al., 2010 |
| ITS3: GCATCGATGAAGAACGCAGC 26SE: TAGAATTCCCCGGTTCGCTCGCCGTTAC | 94°C 4 min; 94°C 30 s, 57°C 45 s, 72°C 45 s, 35 cycles; 72°C 10 min | White et al., 1990; Sun et al., 1994 |
| Barcodes | Amplification efficiency (%) | Success rate of sequencing (%) | Effective sequence ratio (%) |
|---|---|---|---|
| rbcL | 100.0 | 100.0 | 100.0 |
| matK | 100.0 | 100.0 | 100.0 |
| trnL-trnF | 100.0 | 100.0 | 100.0 |
| ITS2 | 100.0 | 89.5 | 89.5 |
表3 条形码扩增和测序成功率
Table 3 The success rate of PCR amplification and sequencing
| Barcodes | Amplification efficiency (%) | Success rate of sequencing (%) | Effective sequence ratio (%) |
|---|---|---|---|
| rbcL | 100.0 | 100.0 | 100.0 |
| matK | 100.0 | 100.0 | 100.0 |
| trnL-trnF | 100.0 | 100.0 | 100.0 |
| ITS2 | 100.0 | 89.5 | 89.5 |
| Barcodes | No. of sequences | Aligned length (bp) | GC content (%) | No. of variable sites | Intraspecific distance (mean) | Interspecific distance (mean) |
|---|---|---|---|---|---|---|
| rbcL | 259 | 684 | 46.0 | 23 | 0.0006 | 0.0161 |
| matK | 207 | 969 | 35.8 | 27 | 0.0032 | 0.0273 |
| trnL-trnF | 365 | 454 | 27.2 | 46 | 0.0041 | 0.0273 |
| ITS2 | 68 | 473 | 72.5 | 51 | 0.0017 | 0.1311 |
表4 序列特征信息
Table 4 Information of sequences characteristic
| Barcodes | No. of sequences | Aligned length (bp) | GC content (%) | No. of variable sites | Intraspecific distance (mean) | Interspecific distance (mean) |
|---|---|---|---|---|---|---|
| rbcL | 259 | 684 | 46.0 | 23 | 0.0006 | 0.0161 |
| matK | 207 | 969 | 35.8 | 27 | 0.0032 | 0.0273 |
| trnL-trnF | 365 | 454 | 27.2 | 46 | 0.0041 | 0.0273 |
| ITS2 | 68 | 473 | 72.5 | 51 | 0.0017 | 0.1311 |
| W+ | W- | Intra relative ranks | N | P value | Result |
|---|---|---|---|---|---|
| trnL-trnF | rbcL | W+=1258, W-=120 | 96 | 2.20E-07 | P<0.05, trnL-trnF>rbcL |
| trnL-trnF | matK | W+=298, W-=297 | 43 | 9.93E-01 | P>0.05, matK=trnL-trnF |
| matK | rbcL | W+=408, W-=57 | 44 | 9.18E-04 | P<0.05, matK>rbcL |
表5 叶绿体条形码片段在龙脑香科物种种内变异Wilcoxon检验
Table 5 Wilcoxon signed rank tests of intraspecific divergence among chloroplast markers in the species of Dipterocarpaceae
| W+ | W- | Intra relative ranks | N | P value | Result |
|---|---|---|---|---|---|
| trnL-trnF | rbcL | W+=1258, W-=120 | 96 | 2.20E-07 | P<0.05, trnL-trnF>rbcL |
| trnL-trnF | matK | W+=298, W-=297 | 43 | 9.93E-01 | P>0.05, matK=trnL-trnF |
| matK | rbcL | W+=408, W-=57 | 44 | 9.18E-04 | P<0.05, matK>rbcL |
| W+ | W- | Inter relative ranks | N | P value | Result |
|---|---|---|---|---|---|
| trnL-trnF | rbcL | W+=8136415.5, W-=1811614.5 | 4560 | 3.78E-296 | P<0.05, trnL-trnF>rbcL |
| trnL-trnF | matK | W+=236579.5, W-=156361.5 | 903 | 1.41E-07 | P<0.05, trnL-trnF>matK |
| matK | rbcL | W+=411099, W-=36832 | 946 | 1.00E-13 | P<0.05, matK>rbcL |
表6 叶绿体条形码片段在龙脑香科物种种间变异Wilcoxon检验
Table 6 Wilcoxon signed rank tests of interspecific divergence among chloroplast markers in the species of Dipterocarpaceae
| W+ | W- | Inter relative ranks | N | P value | Result |
|---|---|---|---|---|---|
| trnL-trnF | rbcL | W+=8136415.5, W-=1811614.5 | 4560 | 3.78E-296 | P<0.05, trnL-trnF>rbcL |
| trnL-trnF | matK | W+=236579.5, W-=156361.5 | 903 | 1.41E-07 | P<0.05, trnL-trnF>matK |
| matK | rbcL | W+=411099, W-=36832 | 946 | 1.00E-13 | P<0.05, matK>rbcL |
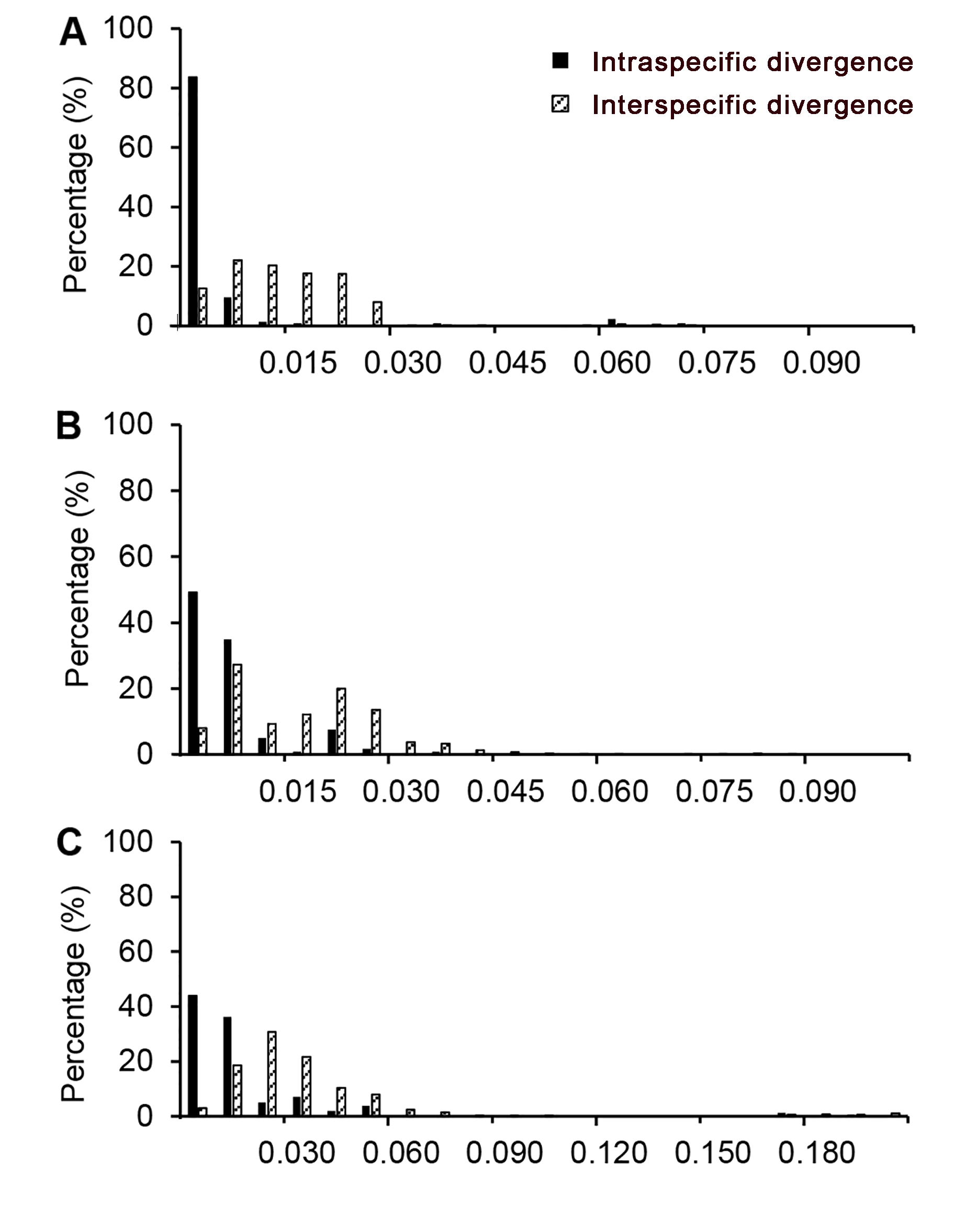
图1 条形码片段在龙脑香科物种种内和种间遗传变异分布(A) rbcL序列; (B) matK序列; (C) trnL-trnF序列
Figure 1 Distribution for intra and inter-specific variation of markers in the species of Dipterocarpaceae(A) rbcL sequence; (B) matK sequence; (C) trnL-trnF sequence
| [1] | 成俊卿 (1980). 中国热带及亚热带木材——识别、材性和利用. 北京: 科学出版社. pp. 413-418. |
| [2] | 戴文君, 周磊, 杨梅 ( 2017). 中国龙脑香科植物研究及利用现状. 世界林业研究 30(6), 46-51. |
| [3] | 李德铢, 曾春霞 ( 2015). 植物DNA条形码研究展望. 生物多样性 23, 297-298. |
| [4] | 李锡文, 李捷 , Ashton P ( 2002). 中国龙脑香料植物纪要. 云南植物研究 24, 409-420. |
| [5] | 卢孟孟, 慈秀芹, 杨国平, 李捷 ( 2013). 亚热带森林乔木树种DNA条形码研究——以哀牢山自然保护区为例. 植物分类与资源学报 35, 733-741. |
| [6] | 罗良才 ( 1988). 云南龙脑香科树种木材识别与利用的研究. 云南林业科技 ( 1), 1-13. |
| [7] | 罗良才 ( 2008). 云南进口龙脑香科树种的木材识别与利用. 西部林业科学 37, 99-107. |
| [8] | 乔梦吉, 陈柏旭, 符韵林 ( 2018). 基于DNA条形码技术的楠属和润楠属5种木材的识别. 基因组学与应用生物学 37, 4013-4021. |
| [9] | 任保青, 陈之端 ( 2010). 植物DNA条形码技术. 植物学报 45, 1-12. |
| [10] | 宋慧芳, 刘海双, 杨义明, 范书田, 李昌禹, 艾军 ( 2017). 山葡萄种质资源DNA条形码通用序列的筛选. 植物学报 52, 723-732. |
| [11] | 童绍全, 陶国达 ( 1990). 龙脑香科. 见: 钱崇澍, 陈焕镛, 主编. 中国植物志, 第50卷2分册. 北京: 科学出版社. pp. 113-131. |
| [12] | 王锦亮, 丁靖垲, 程治英, 杨崇仁 ( 1992). 两种云南龙脑香属植物树脂精油的倍半萜成分及其季节性变化. 云南植物研究 14, 337-342. |
| [13] | 杨家驹, 刘鹏, 卢鸿俊 ( 1989). 龙脑香亚科木材. 木材工业 3(3), 43-49. |
| [14] | 杨家驹, 刘鹏, 卢鸿俊 ( 1991). 娑罗双属商品材. 木材工业 5, 45-50, 55. |
| [15] | 于永福 ( 1999). 中国野生植物保护工作的里程碑——《国家重点保护野生植物名录(第一批)》出台. 植物杂志 ( 5), 3. |
| [16] | 周亮, 黄建平, 黄自云 ( 2013). 热带植物云南娑罗双. 园林 ( 6), 62-63. |
| [17] | 朱华, 王洪 ( 1992). 西双版纳龙脑香科植物纪要. 云南植物研究 14, 21-26. |
| [18] | Ashton PS ( 1982). Flora Malesiana. Series I—Spermatophyta. Flowering Plants, Vol. 9, Part 2. Dipterocarpaceae. The Netherlands: Martinun Nijhoff Pub. |
| [19] | CBOL Plant Working Group, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim KJ, Kress WJ, Schneider H, van AlphenStahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim YD, Lahaye R, Lee HL, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park CW, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi DK, Little DP ( 2009). A DNA barcode for land plants. Proc Natl Acad Sci USA 106, 12794-12797. |
| [20] | Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M ( 2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295-299. |
| [21] | Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C ( 2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5, e8613. |
| [22] | China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW ( 2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108, 19641-19646. |
| [23] | Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW ( 2002). Molecular phylogenetics of Caryo- phyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89, 132-144. |
| [24] | de Boer HJ, Ghorbani A, Manzanilla V, Raclariu AC, Kreziou A, Ounjai S, Osathanunkul M, Gravendeel B ( 2017). DNA metabarcoding of orchid-derived products reveals widespread illegal orchid trade. Proc Roy Soc B 284, 20171182. |
| [25] | Dormontt EE, Boner M, Braun B, Breulmann G, Degen B, Espinoza E, Gardner S, Guillery P, Hermanson JC, Koch G, Lee SL, Kanashiro M, Rimbawanto A, Thomas D, Wiedenhoeft AC, Yin YF, Zahnen J, Lowe AJ ( 2015). Forensic timber identification: it's time to integrate disciplines to combat illegal logging. Biol Conserv 191, 790-798. |
| [26] | Doyle JJ, Doyle JL ( 1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [27] | Fay MF, Cameron KM, Prance GT, Lledó MD, Chase MW ( 1997). Familial relationships of Rhabdodendron( Rhabdodendraceae): plastid rbcL sequences indicate a caryo- phyllid placement. Kew Bull 52, 923-932. |
| [28] | Gasson P, Miller R, Stekel DJ, Whinder F, Ziemińska K ( 2010). Wood identification of Dalbergia nigra( CITES Appendix I) using quantitative wood anatomy, principal components analysis and naïve Bayes classification. Ann Bot 105, 45-56. |
| [29] | Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, Roger A, Thébaud C, Chave J ( 2009). Iden- tification of Amazonian trees with DNA barcodes. PLoS One 4, e7483. |
| [30] | Hassold S, Lowry II PP, Bauert MR, Razafintsalama A, Ramamonjisoa L, Widmer A ( 2016). DNA barcoding of Malagasy rosewoods: towards a molecular identification of CITES-listed Dalbergia species. PLoS One 11, e0157881. |
| [31] | Hebert PDN, Cywinska A, Ball SL, deWaard JR ( 2003). Biological identifications through DNA barcodes. Proc Roy Sci B 270, 313-321. |
| [32] | Heckenhauer J, Samuel R, Ashton PS, Turner B, Barfuss MHJ, Jang TS, Temsch EM, Mccann J, Salim KA, Attanayake AMAS, Chase MW ( 2017). Phylogenetic analyses of plastid DNA suggest a different interpretation of morphological evolution than those used as the basis for previous classifications of Dipterocarpaceae (Malvales). Bot J Linn Soc 185, 1-26. |
| [33] | Hollingsworth PM, Li DZ, van der Bank M, Twyford AD ( 2016). Telling plant species apart with DNA: from barcodes to genomes. Philos Trans Roy Soc B 371, 2015-0338. |
| [34] | Huang XC, Ci XQ, Conran JG, Li J ( 2015). Application of DNA barcodes in Asian tropical trees—a case study from Xishuangbanna Nature Reserve, Southwest China. PLoS One 10, e0129295. |
| [35] | Jiao LC, Yu M, Wiedenhoeft AC, He T, Li JN, Liu B, Jiang XM, Yin YF ( 2018). DNA barcode authentication and library development for the wood of six commercial Pterocarpus species: the critical role of xylarium specimens. Sci Rep 8, 1945. |
| [36] | Kamiya K, Harada K, Tachida H, Ashton PS ( 2005). Phylogeny of PgiC gene in Shorea and its closely related genera (Dipterocarpaceae), the dominant trees in southeast Asian tropical rain forests. Am J Bot 92, 775-788. |
| [37] | Kress WJ, Erickson DL ( 2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2, e508. |
| [38] | Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH ( 2005). Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102, 8369-8374. |
| [39] | Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V ( 2008). DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105, 2923-2928. |
| [40] | Meier R, Shiyang K, Vaidya G, Ng PKL ( 2006). DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55, 715-728. |
| [41] | Newmaster SG, Fazekas AJ, Ragupathy S ( 2006). DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot 84, 335-341. |
| [42] | Ng CH, Lee SL, Tnah LH, Ng KKS, Lee CT, Maria M ( 2016). Genome size variation and evolution in Dipterocarpaceae. Plant Ecol Divers 9, 437-446. |
| [43] | Olmstead RG, Michaels HJ, Scott KM, Palmer JD ( 1992). Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann Mo Bot Gard 79, 249-265. |
| [44] | Schultz J, Wolf M ( 2009). ITS2 sequence-structure analysis in phylogenetics: a how-to manual for molecular syste- matics. Mol Phylogenet Evol 52, 520-523. |
| [45] | Sun Y, Skinner DZ, Liang GH, Hulbert SH ( 1994). Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor Appl Genet 89, 26-32. |
| [46] | Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E ( 2007). Power and limitations of the chloroplast trnL( UAA) intron for plant DNA barcoding. Nucleic Acids Res 35, e14. |
| [47] | Taberlet P, Gielly L, Pautou G, Bouvet J ( 1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17, 1105-1109. |
| [48] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S ( 2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30, 2725-2729. |
| [49] | Trang NTP, Duc NM, Sinh NV, Triest L ( 2015). Application of DNA barcoding markers to the identification of Hopea species. Genet Mol Res 14, 9181-9190. |
| [50] | Tsumura Y, Kado T, Yoshida K, Abe H, Ohtani M, Taguchi Y, Fukue Y, Tani N, Ueno S, Yoshimura K, Kamiya K, Harada K, Takeuchi Y, Diway B, Finkeldey R, Na’iem M, Indrioko S, Ng KKS, Muhammad N, Lee SL ( 2011). Molecular database for classifying Shorea species (Dipterocarpaceae) and techniques for checking the legitimacy of timber and wood products. J Plant Res 124, 35-48. |
| [51] | White TJ, Bruns TD, Lee S, Taylor J ( 1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR Protocols: a Guide to Methods and Applications. San Diego: Academic Press. pp. 315-322. |
| [52] | Wilson EO ( 2016). Half-Earth: Our Planet’s Fight for Life. New York: Liveright Publishing Corporation. |
| [53] | Yu M, Jiao LC, Guo J, Wiedenhoeft AC, He T, Jiang XM, Yin YF ( 2017). DNA barcoding of vouchered xylarium wood specimens of nine endangered Dalbergia species. Planta 246, 1165-1176. |
| [54] | Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB ( 2018). Genome skimming herbarium spe- cimens for DNA barcoding and phylogenomics. Plant Methods 14, 43. |
| [55] | Zhu RW, Li YC, Zhong DL, Zhang JQ ( 2017). Establishment of the most comprehensive ITS2 barcode database to date of the traditional medicinal plant Rhodiola ( Crassulaceae). Sci Rep 7, 10051. |
| [1] | 罗小燕, 李强, 黄晓磊. 戴云山国家级自然保护区访花昆虫DNA条形码数据集[J]. 生物多样性, 2023, 31(8): 23236-. |
| [2] | 邢超, 林依, 周智强, 赵联军, 蒋仕伟, 林蓁蓁, 徐基良, 詹祥江. 基于DNA条形码技术构建王朗国家级自然保护区陆生脊椎动物遗传资源数据库及物种鉴定[J]. 生物多样性, 2023, 31(7): 22661-. |
| [3] | 王露露, 杨智, 杨永. 利用标本组学推进植物超级DNA条形码研究[J]. 植物学报, 2023, 58(5): 831-842. |
| [4] | 吴帆, 刘深云, 江虎强, 王茜, 陈开威, 李红亮. 中华蜜蜂和意大利蜜蜂秋冬期传粉植物多样性比较[J]. 生物多样性, 2023, 31(5): 22528-. |
| [5] | 俞正森, 宋娜, 本村浩之, 高天翔. 中国银口天竺鲷属鱼类的分类厘定[J]. 生物多样性, 2021, 29(7): 971-979. |
| [6] | 肖文宏, 周青松, 朱朝东, 吴东辉, 肖治术. 野生动物监测技术和方法应用进展与展望[J]. 植物生态学报, 2020, 44(4): 409-417. |
| [7] | 胡芮, 王儒晓, 杜诗雨, 李萌, 邢雨辉, 潘达, 徐海根, 孙红英. 扬州宝应湖底栖大型无脊椎动物的生物多样性及其变化[J]. 生物多样性, 2020, 28(12): 1558-1569. |
| [8] | 邵昕宁, 宋大昭, 黄巧雯, 李晟, 姚蒙. |
| [9] | 刘山林. DNA条形码参考数据集构建和序列分析相关的新兴技术[J]. 生物多样性, 2019, 27(5): 526-533. |
| [10] | 舒江平, 罗俊杰, 韦宏金, 严岳鸿. 基于模式产地的分子证据澄清南平鳞毛蕨的分类学地位[J]. 植物学报, 2018, 53(6): 793-800. |
| [11] | 侯勤曦, 慈秀芹, 刘志芳, 徐武美, 李捷. 基于DNA条形码评估西双版纳国家级自然保护区对樟科植物进化历史的保护[J]. 生物多样性, 2018, 26(3): 217-228. |
| [12] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| [13] | 宋慧芳, 刘海双, 杨义明, 范书田, 李昌禹, 艾军. 山葡萄种质资源DNA条形码通用序列的筛选[J]. 植物学报, 2017, 52(6): 723-732. |
| [14] | 郝金凤, 张晓红, 王昱淞, 刘金林, 智永超, 李新江. 白洋淀湿地蝗虫多样性调查及DNA条形码应用研究[J]. 生物多样性, 2017, 25(4): 409-417. |
| [15] | 慈秀芹, 李捷. 系统发育多样性在植物区系研究与 生物多样性保护中的应用[J]. 生物多样性, 2017, 25(2): 175-181. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||