

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 328-334.DOI: 10.11983/CBB18117 cstr: 32102.14.CBB18117
程广前1,†,贾克利1,2,†,李娜1,邓传良1,李书粉1,高武军1,*
收稿日期:2018-05-11
接受日期:2018-08-06
出版日期:2019-05-01
发布日期:2019-11-24
通讯作者:
高武军
基金资助:Guangqian Cheng1,†,Keli Jia1,2,†,Na Li1,Chuanliang Deng1,Shufen Li1,Wujun Gao1,*
Received:2018-05-11
Accepted:2018-08-06
Online:2019-05-01
Published:2019-11-24
Contact:
Wujun Gao
摘要: 在植物基因组中, 叶绿体DNA (cpDNA)序列可以向核基因组转移成为核质体DNA (NUPT)。NUPTs在植物染色体(包括性染色体)的演化过程中具有重要作用, 但目前相关研究比较缺乏。以雌雄异株植物石刁柏(Asparagus officinalis)为材料, 采用生物信息学方法对其核基因组NUPTs进行注释及分析, 并选取叶绿体基因组反向重复区(IR) 2个片段进行染色体定位。结果表明, 石刁柏核基因组中有2 239个NUPTs序列的插入, 总长度为565 970 bp, 占核基因组的0.047%。不同染色体上插入的NUPTs数量存在较大差异, Y染色体上的NUPTs数量、密度及总长度均高于其它染色体, 表明NUPTs在石刁柏性(Y)染色体上累积的更多。石刁柏叶绿体基因组中的IR区、大单拷贝区(LSC)和小单拷贝区(SSC)序列均能够向核基因组转移, 但IR区序列转移频率更高。此外, 对2个IR区的叶绿体序列进行荧光原位杂交, 其中AocpIR1主要分布在所有染色体的着丝粒部位, 而AocpIR2特异性分布在Y染色体上。研究结果为深入揭示石刁柏基因组的结构及其性染色体的演化奠定了坚实的基础。
程广前,贾克利,李娜,邓传良,李书粉,高武军. 石刁柏核质体DNA的生物信息学分析及染色体定位. 植物学报, 2019, 54(3): 328-334.
Guangqian Cheng,Keli Jia,Na Li,Chuanliang Deng,Shufen Li,Wujun Gao. Bioinformatics Analysis and Chromosome Location of Nuclear Integrants of Plastid DNA in Asparagus officinalis. Chinese Bulletin of Botany, 2019, 54(3): 328-334.
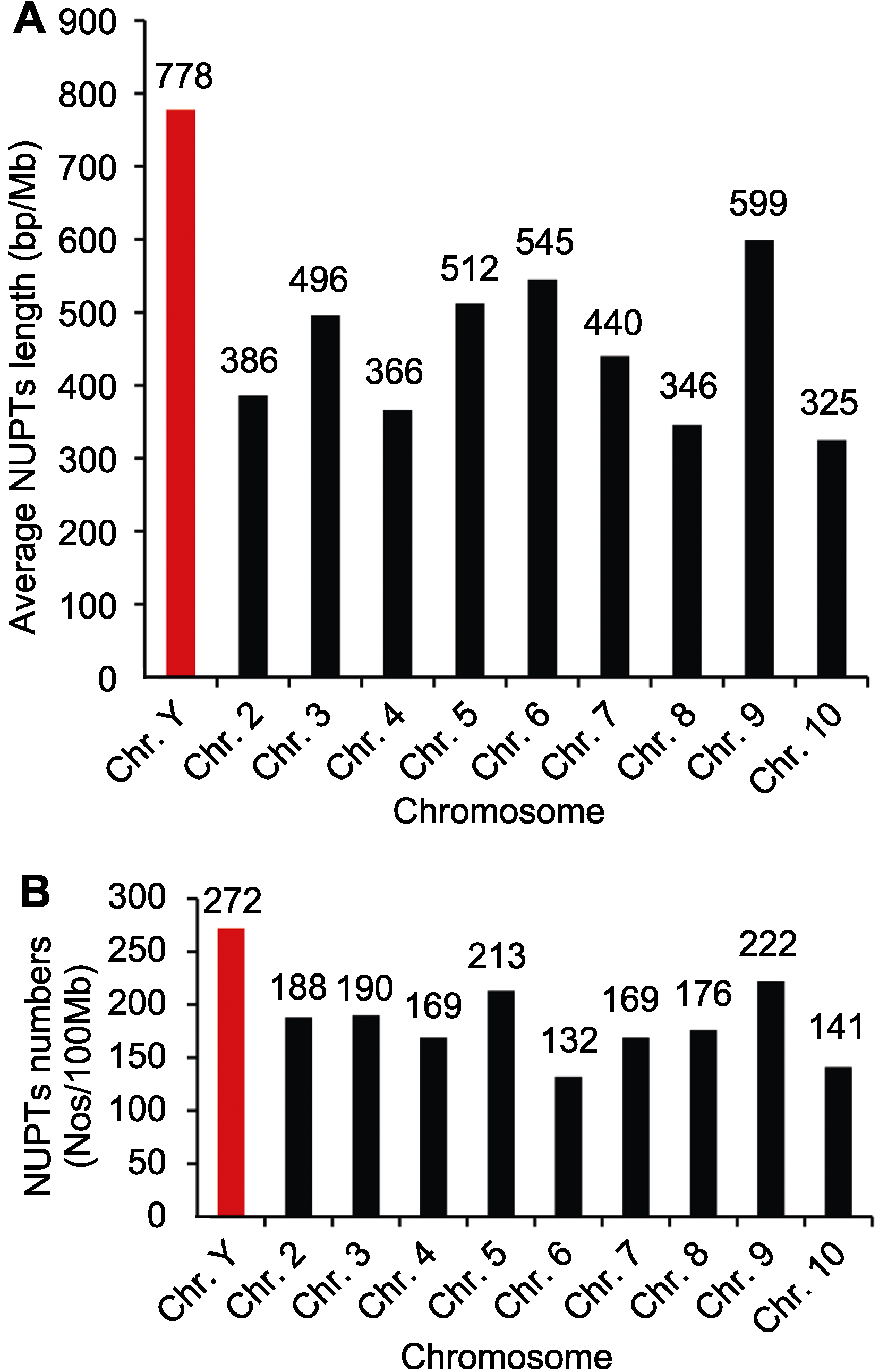
图1 石刁柏不同染色体核质体DNA (NUPTs)长度(A)和数目(B)
Figure 1 The length (A) and number (B) of nuclear integrants of plastid DNAs (NUPTs) in different chromosomes of Asparagus officinalis
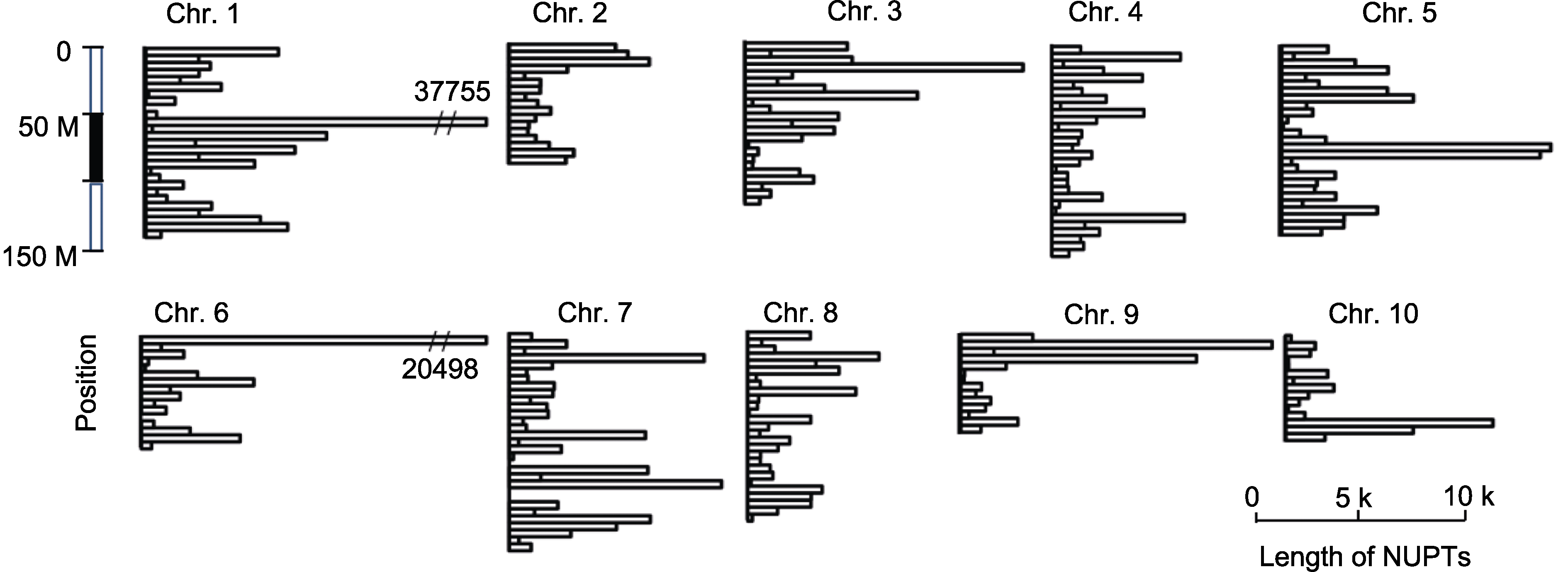
图2 石刁柏基因组核质体DNA (NUPTs)在染色体上的分布模式
Figure 2 The chromosome distribution pattern of nuclear integrants of plastid DNAs (NUPTs) of Asparagus officinalis genome
| IR | LSC | SSC | |
|---|---|---|---|
| Total sequence length (bp) | 26471 | 83821 | 17904 |
| Number of NUPTs | 726 | 1227 | 286 |
| Average density (No./kb) | 27.9 | 14.6 | 15.9 |
| Average length of NUPTs (bp) | 241 | 258 | 246 |
| The length of NUPTs (bp) | 175215 | 316626 | 70394 |
表1 石刁柏核基因组核质体DNA (NUPTs)的叶绿体基因组来源分析
Table 1 Origin analysis of nuclear integrants of plastid DNAs (NUPTs) sequences on the chloroplast genome of Asparagus officinalis
| IR | LSC | SSC | |
|---|---|---|---|
| Total sequence length (bp) | 26471 | 83821 | 17904 |
| Number of NUPTs | 726 | 1227 | 286 |
| Average density (No./kb) | 27.9 | 14.6 | 15.9 |
| Average length of NUPTs (bp) | 241 | 258 | 246 |
| The length of NUPTs (bp) | 175215 | 316626 | 70394 |
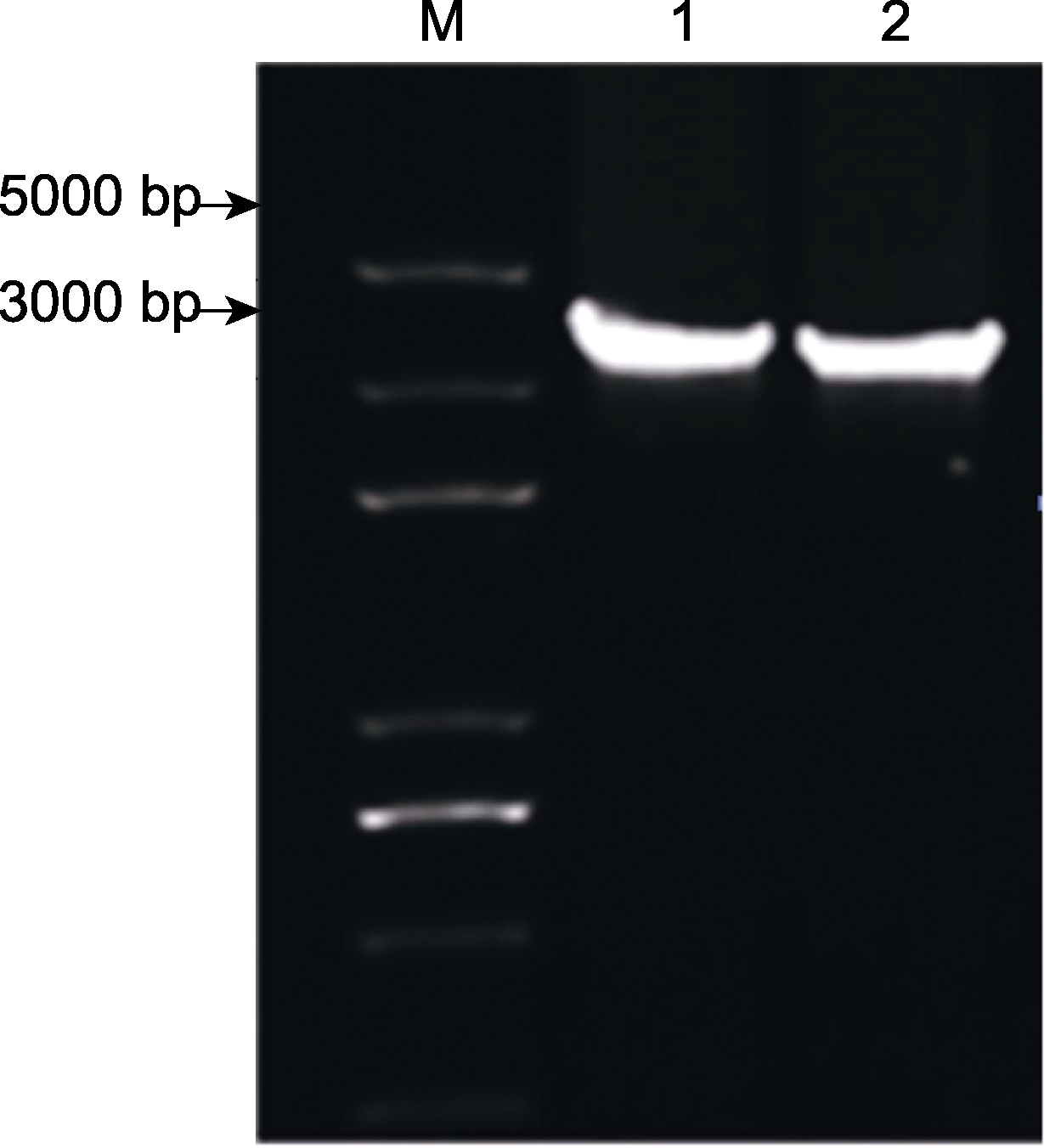
图3 石刁柏2个叶绿体反向重复区(IR)序列的PCR扩增1, 2分别代表AocplR1和AocplR2的扩增结果。
Figure 3 PCR amplification of inverted repeat region (IR) two sequences of region in Asparagus officinalis 1, 2 respresents the amplification results of AocplR1 and AocplR2, respectively.
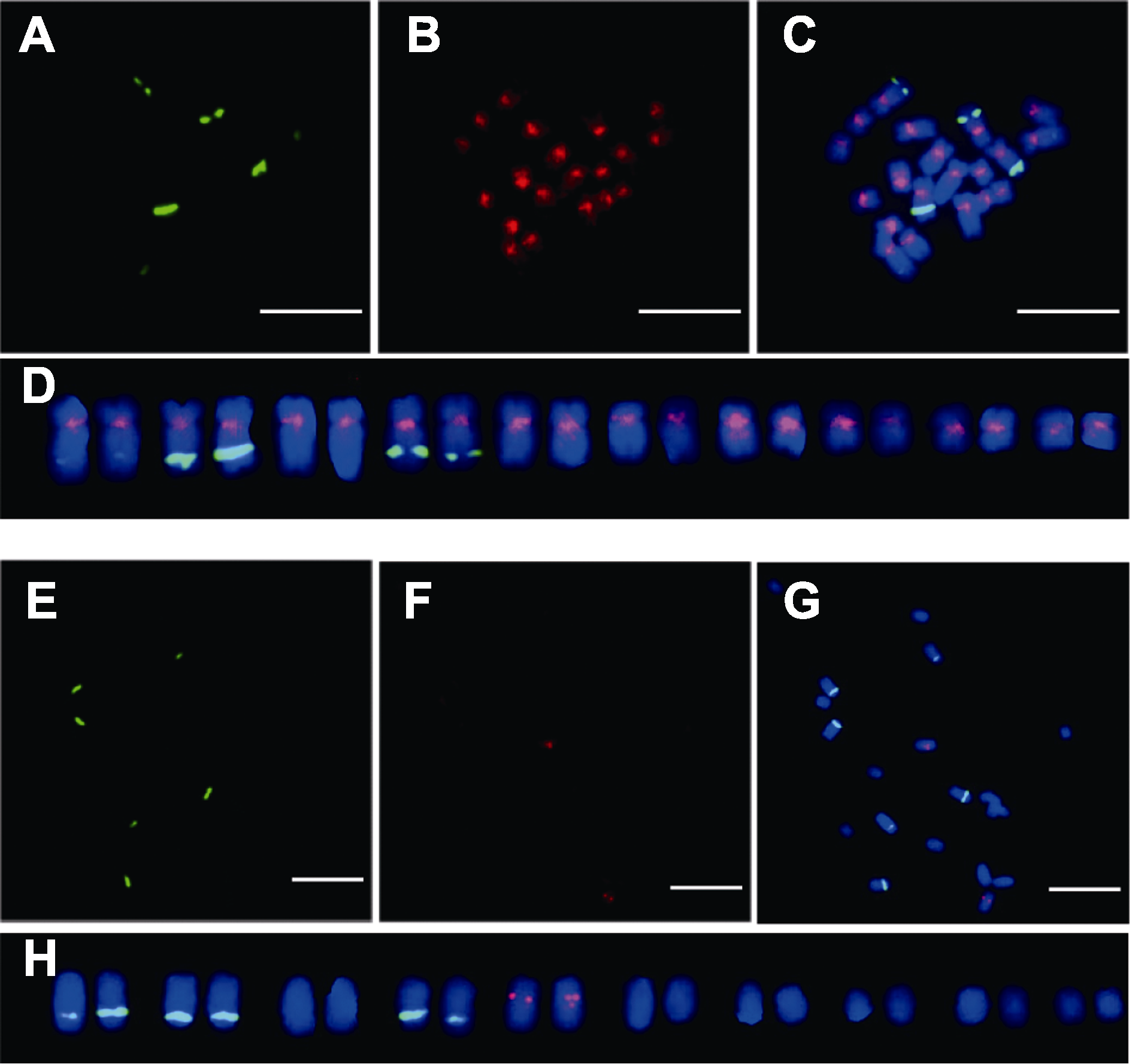
图4 AocpIR1和AocpIR2在石刁柏有丝分裂中期染色体上的定位(A), (E) 45S rDNA杂交信号图; (B), (F) AocpIR1和AocpIR2杂交信号图; (C), (G) 合成图; (D), (H) 核型图。Bars=10 μm
Figure 4 Mitosis metaphase chromosome locations of AocpIR1 and AocpIR2 of Asparagus officinalis (A), (E) FISH result of 45S rDNA; (B), (F) FISH result of AocpIR1 and AocpIR2; (C), (G) Merged picture; (D), (H) Karyotype analysis. Bars=10 μm
| [1] |
高东迎, 何冰, 孙立华 ( 2007). 水稻转座子研究进展. 植物学通报 24, 667-676.
DOI URL |
| [2] |
李巧丽, 延娜, 宋琼, 郭军战 ( 2018). 鲁桑叶绿体基因组序列及特征分析. 植物学报 53, 94-103.
DOI URL |
| [3] |
李书粉, 李旭, 王冰肖, 袁金红, 邓传良, 高武军 ( 2016). 石刁柏雄性偏向核质体DNA的克隆与分析. 西北植物学报 36, 2385-2390.
DOI URL |
| [4] |
Abbott JK, Nordén AK, Hansson B ( 2017). Sex chromosome evolution: historical insights and future perspectives. Proc Roy Soc B 284, 20162806.
DOI URL |
| [5] |
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ ( 1990). Basic local alignment search tool. J Mol Biol 215, 403-410.
DOI URL |
| [6] | Doyle JJ, Doyle JL ( 1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [7] |
Feschotte C, Pritham EJ ( 2007). DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41, 331-368.
DOI URL PMID |
| [8] |
Guo XY, Ruan SL, Hu WM, Cai DG, Fan LJ ( 2008). Chloroplast DNA insertions into the nuclear genome of rice: the genes, sites and ages of insertion involved. Funct Integr Genomics 8, 101-108.
DOI URL |
| [9] |
Harkess A, Zhou JS, Xu CY, Bowers JE, van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A, Tang HB, Ray J, Groenendijk J, Arikit S, Mathioni SM, Nakano M, Shan HY, Telgmann-Rauber A, Kanno A, Yue Z, Chen HX, Li WQ, Chen YL, Xu XY, Zhang YP, Luo SC, Chen HL, Gao JM, Mao ZC, Pires JC, Luo MZ, Kudrna D, Wing RA, Meyers BC, Yi KX, Kong HZ, Lavrijsen P, Sunseri F, Falavigna A, Ye Y, Leebens-Mack JH, Chen GY ( 2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun 8, 1279.
DOI URL PMID |
| [10] |
Kejnovsky E, Hobza R, Cermak T, Kubat Z, Vyskot B ( 2009). The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 102, 533-541.
DOI |
| [11] |
Kejnovsky E, Kubat Z, Hobza R, Lengerova M, Sato S, Tabata S, Fukui K, Matsunaga S, Vyskot B ( 2006). Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica 128, 167-175.
DOI URL PMID |
| [12] |
Kleine T, Maier UG, Leister D ( 2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60, 115-138.
DOI URL PMID |
| [13] |
Li SF, Zhang GJ, Yuan JH, Deng CL, Gao WJ ( 2016). Repetitive sequences and epigenetic modification: inseparable partners play important roles in the evolution of plant sex chromosomes. Planta 243, 1083-1095.
DOI URL |
| [14] | Löptien H ( 1979). Identification of the sex chromosome pair in asparagus ( Asparagus officinalis L.). Z Pflanzenzüecht 82, 162-173. |
| [15] |
Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, Schmutzer T, Scholz U, Gundlach H, Wicker T, Šimková H, Novák P, Neumann P, Kubaláková M, Bauer E, Haseneyer G, Fuchs J, Doležel J, Stein N, Mayer KFX, Houben A ( 2012). Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci USA 109, 13343-13346.
DOI URL |
| [16] |
Matsuo M, Ito Y, Yamauchi R, Obokata J ( 2005). The rice nuclear genome continuously integrates, shuffles, and eli- minates the chloroplast genome to cause chloroplast- nuclear DNA flux. Plant Cell 17, 665-675.
DOI URL |
| [17] |
Michalovova M, Vyskot B, Kejnovsky E ( 2013). Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity 111, 314-320.
DOI URL PMID |
| [18] |
Richly E, Leister D ( 2004). NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21, 1081-1084.
DOI URL PMID |
| [19] | Sheng WT, Chai XW, Rao YS, Tu XT, Du SG ( 2017). Complete chloroplast genome sequence of asparagus ( Asparagus officinalis L.) and its phylogenetic position within asparagales. J Plant Breed Genet 5, 121-128. |
| [20] | Steflova P, Hobza R, Vyskot B, Kejnovsky E ( 2014). Strong accumulation of chloroplast DNA in the Y chromosomes of Rumex acetosa and Silene latifolia. Cytogenet Genome Res 142, 59-65. |
| [21] | Timmis JN, Ayliffe MA, Huang CY, Martin W ( 2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5, 123-135. |
| [22] |
VanBuren R, Ming R ( 2013). Organelle DNA accumulation in the recently evolved papaya sex chromosomes. Mol Genet Genomics 288, 277-284.
DOI URL |
| [23] |
Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM ( 2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol 8, 36.
DOI URL PMID |
| [24] |
Yoshida T, Furihata HY, Kawabe A ( 2014). Patterns of genomic integration of nuclear chloroplast DNA fragments in plant species. DNA Res 21, 127-140.
DOI URL PMID |
| [1] | 罗兰莎, 宋雯佩, 化青珠, 李大卫, 梁红, 张宪智. 植物性别决定基因及其表观遗传调控研究进展[J]. 植物学报, 2024, 59(2): 278-290. |
| [2] | 彭丹, 武志强. 植物雌雄异株性别决定研究进展[J]. 生物多样性, 2022, 30(3): 21416-. |
| [3] | 李格,孟小庆,李宗芸,朱明库. 甘薯盐胁迫响应基因IbMYB3的表达特征及生物信息学分析[J]. 植物学报, 2020, 55(1): 38-48. |
| [4] | 秦力, 陈景丽, 潘长田, 叶蕾, 卢钢. 植物性染色体进化及性别决定基因研究进展[J]. 植物学报, 2016, 51(6): 841-848. |
| [5] | 汪丽虹 王星 崔凯荣 焦成瑾 王亚馥. 石刁柏及党参体细胞胚发生中的淀粉代谢动态[J]. 植物学报, 1996, 13(01): 41-45. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||