

Research Progress on Plant Sex-determination Genes and Their Epigenetic Regulation
Received date: 2023-07-03
Accepted date: 2023-12-19
Online published: 2023-12-21
The mechanism of sex determination in dioecious plants is a hot topic in reproductive biology, evolution and ecology, and so on. During recent years, the sex determination genes of several important crops, such as asparagus, kiwifruit, and poplar, have been revealed and applied to produce sex-specific products. New germplasms with bisexual flowers have also been developed for molecular breeding. In this review, the genetic bases of sex determination were systematically analyzed from the aspects of sex chromosomes and sex-determination genes in dioecious plants. Subsequently, the epigenetic regulatory roles of non-coding RNA and DNA methylation in plant sex determination were discussed. Finally, it was suggested that further comparative studies of sex determination genes among different dioecious plants should be carried out. More efforts are also needed to pay on the epigenetic regulations of plant sex determination. This work will not only facilitate to understand the molecular mechanisms of sex determination in dioecious plants, but also help to expand the application values of sex determination studies.
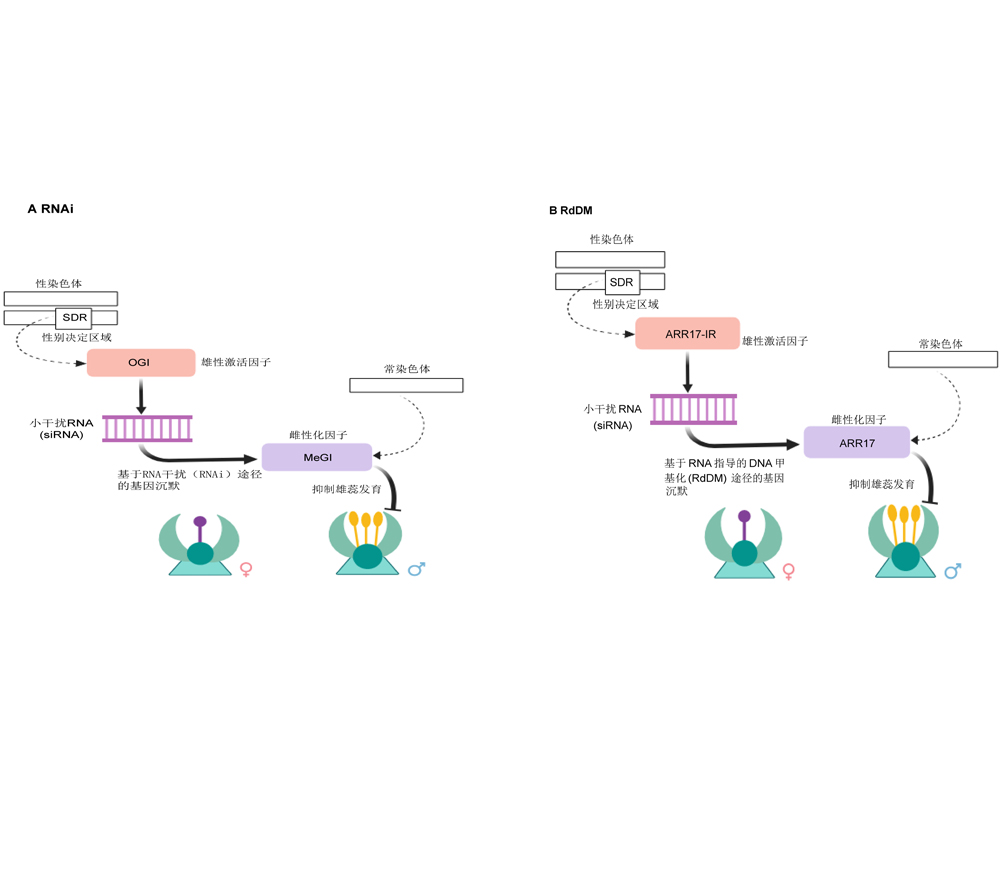
Lansha Luo , Wenpei Song , Qingzhu Hua , Dawei Li , Hong Liang , Xianzhi Zhang . Research Progress on Plant Sex-determination Genes and Their Epigenetic Regulation[J]. Chinese Bulletin of Botany, 2024 , 59(2) : 278 -290 . DOI: 10.11983/CBB23088
| [1] | Akagi T, Henry IM, Kawai T, Comai L, Tao R (2016). Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28, 2905-2915. |
| [2] | Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R (2018). A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30, 780-795. |
| [3] | Akagi T, Henry IM, Tao R, Comai L (2014). A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346, 646-650. |
| [4] | Akagi T, Pilkington SM, Varkonyi-Gasic E, Henry IM, Sugano SS, Sonoda M, Firl A, McNeilage MA, Douglas MJ, Wang TC, Rebstock R, Voogd C, Datson P, Allan AC, Beppu K, Kataoka I, Tao R (2019). Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat Plants 5, 801-809. |
| [5] | Akagi T, Shirasawa K, Nagasaki H, Hirakawa H, Tao R, Comai L, Henry IM (2020). The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet 16, e1008566. |
| [6] | Almeida P, Proux-Wera E, Churcher A, Soler L, Dainat J, Pucholt P, Nordlund J, Martin T, R?nnberg-W?stljung AC, Nystedt B, Berlin S, Mank JE (2020). Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion. BMC Biol 18, 78. |
| [7] | Axtell MJ (2013). Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64, 137-159. |
| [8] | Baránková S, Pascual-Díaz JP, Sultana N, Alonso-Lifante MP, Balant M, Barros K, D'Ambrosio U, Malinská H, Peska V, Lorenzo IP, Kova?ík A, Vyskot B, Janou?ek B, Garcia S (2020). Sex-chrom, a database on plant sex chromosomes. New Phytol 227, 1594-1604. |
| [9] | Cartolano M, Castillo R, Efremova N, Kuckenberg M, Zethof J, Gerats T, Schwarz-Sommer Z, Vandenbussche M (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet 39, 901-905. |
| [10] | Charlesworth B, Charlesworth D (1978). A model for the evolution of dioecy and gynodioecy. Am Nat 112, 975-997. |
| [11] | Cho J, Koo DH, Nam YW, Han CT, Lim HT, Bang JW, Hur Y (2005). Isolation and characterization of cDNA clones expressed under male sex expression conditions in a monoecious cucumber plant (Cucumis sativus L. cv. ‘Winter Long’). Euphytica 146, 271-281. |
| [12] | Cormier F, Lawac F, Maledon E, Gravillon MC, Nudol E, Mournet P, Vignes H, Cha?r H, Arnau G (2019). A reference high-density genetic map of greater yam (Dioscorea alata L.). Theor Appl Genet 132, 1733-1744. |
| [13] | Diggle PK, Di Stilio VS, Gschwend AR, Golenberg EM, Moore RC, Russell JRW, Sinclair JP (2011). Multiple developmental processes underlie sex differentiation in angiosperms. Trends Genet 27, 368-376. |
| [14] | Erdmann RM, Picard CL (2020). RNA-directed DNA methylation. PLoS Genet 16, e1009034. |
| [15] | Fang JB, Zhong CH (2019). Fruit scientific research in New China in the past 70 years: kiwifruit. J Fruit Sci 36, 1352-1359. (in Chinese) |
| 方金豹, 钟彩虹 (2019). 新中国果树科学研究70年——猕猴桃. 果树学报 36, 1352-1359. | |
| [16] | Fraser LG, Tsang GK, Datson PM, De Silva HN, Harvey CF, Gill GP, Crowhurst RN, McNeilage MA (2009). A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics 10, 102. |
| [17] | Fu JQ, Qin BT, Cai JY, Zhang LJ (2022). Research progress of plant sex differentiation. Mol Plant Breed 20, 4833-4839. (in Chinese) |
| 傅靖棋, 秦柏婷, 蔡佳友, 张丽杰 (2022). 植物性别分化研究进展. 分子植物育种 20, 4833-4839. | |
| [18] | Golicz AA, Bhalla PL, Singh MB (2018). lncRNAs in plant and animal sexual reproduction. Trends Plant Sci 23, 195-205. |
| [19] | Govindarajulu R, Liston A, Ashman TL (2013). Sex-determining chromosomes and sexual dimorphism: insights from genetic mapping of sex expression in a natural hybrid Fragaria × ananassa subsp. cuneifolia. Heredity 110, 430-438. |
| [20] | Harkess A, Huang K, van der Hulst R, Tissen B, Caplan JL, Koppula A, Batish M, Meyers BC, Leebens-Mack J (2020). Sex determination by two Y-linked genes in garden asparagus. Plant Cell 32, 1790-1796. |
| [21] | Harkess A, Zhou JS, Xu CY, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A, Tang HB, Ray J, Groenendijk J, Arikit S, Mathioni SM, Nakano M, Shan HY, Telgmann-Rauber A, Kanno A, Yue Z, Chen HX, Li WQ, Chen YL, Xu XY, Zhang YP, Luo SC, Chen HL, Gao JM, Mao ZC, Pires JC, Luo MZ, Kudrna D, Wing RA, Meyers BC, Yi KX, Kong HZ, Lavrijsen P, Sunseri F, Falavigna A, Ye Y, Leebens-Mack JH, Chen GY (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun 8, 1279. |
| [22] | Hobza R, Hudzieczek V, Kubat Z, Cegan R, Vyskot B, Kejnovsky E, Janousek B (2018). Sex and the flower- developmental aspects of sex chromosome evolution. Ann Bot 122, 1085-1101. |
| [23] | Hou J, Ye N, Zhang DF, Chen YN, Fang LC, Dai XG, Yin TM (2015). Different autosomes evolved into sex chromosomes in the sister genera of Salix and Populus. Sci Rep 5, 9076. |
| [24] | Hough J, Hollister JD, Wang W, Barrett SCH, Wright SI (2014). Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci USA 111, 7713-7718. |
| [25] | Hung YH, Slotkin RK (2021). The initiation of RNA interference (RNAi) in plants. Curr Opin Plant Biol 61, 102014. |
| [26] | Hyden B, Zou JZ, Wilkerson DG, Carlson CH, Robles AR, DiFazio SP, Smart LB (2023). Structural variation of a sex-linked region confers monoecy and implicates GATA15 as a master regulator of sex in Salix purpurea. New Phytol 238, 2512-2523. |
| [27] | Iocco-Corena P, Cha?b J, Torregrosa L, Mackenzie D, Thomas MR, Smith HM (2021). VviPLATZ1 is a major factor that controls female flower morphology determination in grapevine. Nat Commun 12, 6995. |
| [28] | Jia HM, Jia HJ, Cai QL, Wang Y, Zhao HB, Yang WF, Wang GY, Li YH, Zhan DL, Shen YT, Niu QF, Chang L, Qiu J, Zhao L, Xie HB, Fu WY, Jin J, Li XW, Jiao Y, Zhou CC, Tu T, Chai CY, Gao JL, Fan LJ, van de Weg E, Wang JY, Gao ZS (2019). The red bayberry genome and genetic basis of sex determination. Plant Biotechnol J 17, 397-409. |
| [29] | K?fer J, Bewick A, Andres-Robin A, Lapetoule G, Harkess A, Ca?us J, Fogliani B, Gateblé G, Ralph P, dePamphilis CW, Picard F, Scutt C, Marais GAB, Leebens-Mack J (2022). A derived ZW chromosome system in Amborella trichopoda, representing the sister lineage to all other extant flowering plants. New Phytol 233, 1636-1642. |
| [30] | Karlov GI, Danilova TV, Horlemann C, Weber G (2003). Molecular cytogenetics in hop (Humulus lupulus L.) and identification of sex chromosomes by DAPI-banding. Euphytica 132, 185-190. |
| [31] | Kazama Y, Kitoh M, Kobayashi T, Ishii K, Krasovec M, Yasui Y, Abe T, Kawano S, Filatov DA (2022). A CLAVATA3-like gene acts as a gynoecium suppression function in white campion. Mol Biol Evol 39, msac195. |
| [32] | Khodaeiaminjan M, Kafkas E, Güeny M, Kafkas S (2017). Development and linkage mapping of novel sex-linked markers for marker-assisted cultivar breeding in pistachio (Pistacia vera L.). Mol Breed 37, 98. |
| [33] | Latrasse D, Rodriguez-Granados NY, Veluchamy A, Mariappan KG, Bevilacqua C, Crapart N, Camps C, Sommard V, Raynaud C, Dogimont C, Boualem A, Benhamed M, Bendahmane A (2017). The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenet Chromatin 10, 22. |
| [34] | Leite Montalv?o AP, Kersten B, Fladung M, Müller NA (2021). The diversity and dynamics of sex determination in dioecious plants. Front Plant Sci 11, 580488. |
| [35] | Leite Montalv?o AP, Kersten B, Kim G, Fladung M, Müller NA (2022). ARR17 controls dioecy in Populus by repressing B-class MADS-box gene expression. Philos Trans Roy Soc B Biol Sci 377, 20210217. |
| [36] | Li CQ (2014). The Research on MiRNA and Bioinformatics Analysis in Carica papaya, Eucalyptus grandis and Other Two Plant Species. Doctoral dissertation. Haikou: Hainan University. pp. 1-237. (in Chinese) |
| 李崇奇 (2014). 番木瓜及巨桉等四种植物miRNA研究和生物信息学分析. 博士论文. 海口: 海南大学. pp. 1-237. | |
| [37] | Li N, Zhou J, Zhang WQ, Liu WJ, Wang BX, She HB, Mirbahar AA, Li SF, Zhang YL, Gao WJ, Qian W, Deng CL (2023a). A rapid method for assembly of single chromosome and identification of sex determination region based on single-chromosome sequencing. New Phytol 240, 892-903. |
| [38] | Li SF, Li S, Deng CL, Lu LD, Gao WJ (2015). Role of transposons in origin and evolution of plant XY sex chromosomes. Hereditas 37, 157-164. (in Chinese) |
| 李书粉, 李莎, 邓传良, 卢龙斗, 高武军 (2015). 转座子在植物XY性染色体起源与演化过程中的作用. 遗传 37, 157-164. | |
| [39] | Li SF, Lv CC, Lan LN, Jiang KL, Zhang YL, Li N, Deng CL, Gao WJ (2021). DNA methylation is involved in sexual differentiation and sex chromosome evolution in the dioecious plant garden asparagus. Hortic Res 8, 198. |
| [40] | Li YL, Wang DY, Wang WW, Yang WL, Gao JW, Zhang WY, Shan LX, Kang MH, Chen Y, Ma T (2023b). A chromosome-level Populus qiongdaoensis genome assembly provides insights into tropical adaptation and a cryptic turnover of sex determination. Mol Ecol 32, 1366-1380. |
| [41] | Liao QG, Du R, Gou JB, Guo LJ, Shen H, Liu HL, Nguyen JK, Ming R, Yin TM, Huang SW, Yan JB (2020). The genomic architecture of the sex-determining region and sex-related metabolic variation in Ginkgo biloba. Plant J 104, 1399-1409. |
| [42] | Liu HL, Wang RH, Mao BG, Zhao BR, Wang JB (2019). Identification of lncRNAs involved in rice ovule development and female gametophyte abortion by genome-wide screening and functional analysis. BMC Genomics 20, 90. |
| [43] | Liu J, Chatham L, Aryal R, Yu QY, Ming R (2018). Differential methylation and expression of HUA1 ortholog in three sex types of papaya. Plant Sci 272, 99-106. |
| [44] | Liu Y, Wang SB, Li LZ, Yang T, Dong SS, Wei T, Wu SD, Liu YB, Gong YQ, Feng XY, Ma JC, Chang GX, Huang JL, Yang Y, Wang HL, Liu M, Xu Y, Liang HP, Yu J, Cai YQ, Zhang ZW, Fan YN, Mu WX, Sahu SK, Liu SC, Lang XA, Yang LL, Li N, Habib S, Yang YQ, Lindstrom AJ, Liang P, Goffinet B, Zaman S, Wegrzyn JL, Li DX, Liu J, Cui J, Sonnenschein EC, Wang XB, Ruan J, Xue JY, Shao ZQ, Song C, Fan GY, Li Z, Zhang LS, Liu JQ, Liu ZJ, Jiao YN, Wang XQ, Wu H, Wang ET, Lisby M, Yang HM, Wang J, Liu X, Xu X, Li N, Soltis PS, Van de Peer Y, Soltis DE, Gong X, Liu H, Zhang SZ (2022). The Cycas genome and the early evolution of seed plants. Nat Plants 8, 389-401. |
| [45] | Liu ZY, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu QY, Pearl HM, Kim MS, Charlton JW, Stiles JI, Zee FT, Paterson AH, Ming R (2004). A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427, 348-352. |
| [46] | Lu J, Chen YN, Yin TM (2021). Research progress on sex determination genes of woody plants. Chin Bull Bot 56, 90-103. (in Chinese) |
| 陆静, 陈赢男, 尹佟明 (2021). 木本植物性别决定基因研究进展. 植物学报 56, 90-103. | |
| [47] | Ma JX, Yan BX, Qu YY, Qin FF, Yang YT, Hao XJ, Yu JJ, Zhao Q, Zhu DY, Ao GM (2008). Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J Cell Biochem 105, 136-146. |
| [48] | Ma XK, Yu LA, Fatima M, Wadlington WH, Hulse-Kemp AM, Zhang XT, Zhang SC, Xu XD, Wang JJ, Huang HX, Lin J, Deng B, Liao ZY, Yang ZH, Ma YH, Tang HB, Van Deynze A, Ming R (2022). The spinach YY genome reveals sex chromosome evolution, domestication, and introgression history of the species. Genome Biol 23, 75. |
| [49] | Marks RA, Smith JJ, Cronk Q, Grassa CJ, McLetchie DN (2019). Genome of the tropical plant Marchantia inflexa: implications for sex chromosome evolution and dehydration tolerance. Sci Rep 9, 8722. |
| [50] | Massonnet M, Cochetel N, Minio A, Vondras AM, Lin J, Muyle A, Garcia JF, Zhou YF, Delledonne M, Riaz S, Figueroa-Balderas R, Gaut BS, Cantu D (2020). The genetic basis of sex determination in grapes. Nat Commun 11, 2902. |
| [51] | McDaniel SF, Neubig KM, Payton AC, Quatrano RS, Cove DJ (2013). Recent gene-capture on the UV sex chromosomes of the moss Ceratodon purpureus. Evolution 67, 2811-2822. |
| [52] | Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, Schneider VA, Potapova T, Wood J, Chow W, Armstrong J, Fredrickson J, Pak E, Tigyi K, Kremitzki M, Markovic C, Maduro V, Dutra A, Bouffard GG, Chang AM, Hansen NF, Wilfert AB, Thibaud-Nissen F, Schmitt AD, Belton JM, Selvaraj S, Dennis MY, Soto DC, Sahasrabudhe R, Kaya G, Quick J, Loman NJ, Holmes N, Loose M, Surti U, Risques RA, Lindsay TAG, Fulton R, Hall I, Paten B, Howe K, Timp W, Young A, Mullikin JC, Pevzner PA, Gerton JL, Sullivan BA, Eichler EE, Phillippy AM (2020). Telomere-to-te- lomere assembly of a complete human X chromosome. Nature 585, 79-84. |
| [53] | Ming R, Bendahmane A, Renner SS (2011). Sex chromosomes in land plants. Annu Rev Plant Biol 62, 485-514. |
| [54] | Montgomery JS, Sadeque A, Giacomini DA, Brown PJ, Tranel PJ (2019). Sex-specific markers for waterhemp (Amaranthus tuberculatus) and palmer amaranth (Amaranthus palmeri). Weed Sci 67, 412-418. |
| [55] | Müller NA, Kersten B, Montalv?o APL, M?hler N, Bernhardsson C, Br?utigam K, Lorenzo ZC, Hoenicka H, Kumar V, Mader M, Pakull B, Robinson KM, Sabatti M, Vettori C, Ingvarsson PK, Cronk Q, Street NR, Fladung M (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat Plants 6, 630-637. |
| [56] | Okada S, Sone T, Fujisawa M, Nakayama S, Takenaka M, Ishizaki K, Kono K, Shimizu-Ueda Y, Hanajiri T, Yamato KT, Fukuzawa H, Brennicke A, Ohyama K (2001). The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc Natl Acad Sci USA 98, 9454-9459. |
| [57] | Pakull B, Groppe K, Mecucci F, Gaudet M, Sabatti M, Fladung M (2011). Genetic mapping of linkage group XIX and identification of sex-linked SSR markers in a Populus tremula × Populus tremuloides cross. Can J For Res 41, 245-253. |
| [58] | Paolucci I, Gaudet M, Jorge V, Beritognolo I, Terzoli S, Kuzminsky E, Muleo R, Mugnozza GS, Sabatti M (2010). Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet Genomes 6, 863-875. |
| [59] | Peng D, Wu ZQ (2022). Progress on sex determination of dioecious plants. Biodiver Sci 30, 21416. (in Chinese) |
| 彭丹, 武志强 (2022). 植物雌雄异株性别决定研究进展. 生物多样性 30, 21416. | |
| [60] | Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, K?fer J, Karlov G, Marais GAB (2020). An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Res 30, 164-172. |
| [61] | Puterova J, Razumova O, Martinek T, Alexandrov O, Divashuk M, Kubat Z, Hobza R, Karlov G, Kejnovsky E (2017). Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biol Evol 9, 197-212. |
| [62] | Qi LQ (2018). Involvement of Cold-induced TaHD2C Gene in Regulating the Floral Transition of Transgenic Arabidopsis. Master's thesis. Jinan: Shandong University. pp. 1-91. (in Chinese) |
| 齐鲁琦 (2018). 冷诱导TaHD2C在拟南芥花期调控中的作用. 硕士论文. 济南: 山东大学. pp. 1-91. | |
| [63] | Qin L, Chen JL, Pan CT, Ye L, Lu G (2016). Research progress in plant sex chromosome evolution and sex determination genes. Chin Bull Bot 51, 841-848. (in Chinese) |
| 秦力, 陈景丽, 潘长田, 叶蕾, 卢钢 (2016). 植物性染色体进化及性别决定基因研究进展. 植物学报 51, 841-848. | |
| [64] | Renner SS (2016). Pathways for making unisexual flowers and unisexual plants: moving beyond the “two mutations linked on one chromosome” model. Am J Bot 103, 587-589. |
| [65] | Rifkin JL, Beaudry FEG, Humphries Z, Choudhury BI, Barrett SCH, Wright SI (2021). Widespread recombination suppression facilitates plant sex chromosome evolution. Mol Biol Evol 38, 1018-1030. |
| [66] | Scharmann M, Grafe TU, Metali F, Widmer A (2019). Sex is determined by XY chromosomes across the radiation of dioecious Nepenthes pitcher plants. Evol Lett 3, 586-597. |
| [67] | Shao FQ, Luo XR, Wang Q, Zhang XZ, Wang WC (2023). Advances in research of DNA methylation regulation during fruit ripening. Acta Hortic Sini 50, 197-208. (in Chinese) |
| 邵凤清, 罗秀荣, 王奇, 张宪智, 王文彩 (2023). 果实成熟过程中的DNA甲基化调控研究进展. 园艺学报 50, 197-208. | |
| [68] | Somyong S, Poopear S, Sunner SK, Wanlayaporn K, Jomchai N, Yoocha T, Ukoskit K, Tangphatsornruang S, Tragoonrung S (2016). ACC oxidase and miRNA- 159a, and their involvement in fresh fruit bunch yield (FFB) via sex ratio determination in oil palm. Mol Genet Genomics 291, 1243-1257. |
| [69] | Tamiru M, Natsume S, Takagi H, White B, Yaegashi H, Shimizu M, Yoshida K, Uemura A, Oikawa K, Abe A, Urasaki N, Matsumura H, Babil P, Yamanaka S, Matsumoto R, Muranaka S, Girma G, Lopez-Montes A, Gedil M, Bhattacharjee R, Abberton M, Kumar P, Rabbi I, Tsujimura M, Terachi T, Haerty W, Corpas M, Kamoun S, Kahl G, Takagi H, Asiedu R, Terauchi R (2017). Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination. BMC Biol 15, 86. |
| [70] | Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C (2007). Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol Genet Genomics 278, 221-234. |
| [71] | Tennessen JA, Wei N, Straub SCK, Govindarajulu R, Liston A, Ashman TL (2018). Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol 16, e2006062. |
| [72] | The Tree of Sex Consortium(2014). Tree of sex: a database of sexual systems. Sci Data 1, 140015. |
| [73] | Torres MF, Mathew LS, Ahmed I, Al-Azwani IK, Krueger R, Rivera-Nu?ez D, Mohamoud YA, Clark AG, Suhre K, Malek JA (2018). Author correction: genus-wide sequencing support a two-locus model for sex-determination in Phoenix. Nat Commun 9, 5219. |
| [74] | Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasj?rvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Lepl? JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596-1604. |
| [75] | Urasaki N, Tarora K, Shudo A, Ueno H, Tamaki M, Miyagi N, Adaniya S, Matsumura H (2012). Digital transcriptome analysis of putative sex-determination genes in papaya (Carica papaya). PLoS One 7, e40904. |
| [76] | Varkonyi-Gasic E, Wang TC, Cooney J, Jeon S, Voogd C, Douglas MJ, Pilkington SM, Akagi T, Allan AC (2021). Shy girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signaling. New Phytol 230, 1461-1475. |
| [77] | Veltsos P, Ridout KE, Toups MA, González-Martínez SC, Muyle A, Emery O, Rastas P, Hudzieczek V, Hobza R, Vyskot B, Marais GAB, Filatov DA, Pannell JR (2019). Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics 212, 815-835. |
| [78] | Wang DY, Li YL, Li MM, Yang WL, Ma XZ, Zhang L, Wang YB, Feng YL, Zhang YY, Zhou R, Sanderson BJ, Keefover-Ring K, Yin TM, Smart LB, DiFazio SP, Liu JQ, Olson M, Ma T (2022). Repeated turnovers keep sex chromosomes young in willows. Genome Biol 23, 200. |
| [79] | Wang Y, Cai XJ, Zhang Y, H?randl E, Zhang ZX, He L (2023). The male-heterogametic sex determination system on chromosome 15 of Salix triandra and Salix arbutifolia reveals ancestral male heterogamety and subsequent turnover events in the genus Salix. Heredity 130, 122-134. |
| [80] | Xue LJ, Wu HT, Chen YN, Li XP, Hou J, Lu J, Wei SY, Dai XG, Olson MS, Liu JQ, Wang MX, Charlesworth D, Yin T (2020). Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat Commun 11, 5893. |
| [81] | Yan MK, Xu Q, Liu CY, Zhang Q, Yao XH (2015). Preliminary investigation on sex differentiation of Actinidia chinensis by high-throughput microRNAs sequencing. Acta Hortic Sini 42, 1260-1272. (in Chinese) |
| 闫明科, 徐强, 刘春燕, 张琼, 姚小洪 (2015). 基于microRNA深度测序的猕猴桃性别分化初探. 园艺学报 42, 1260-1272. | |
| [82] | Yang HW, Akagi T, Kawakatsu T, Tao R (2019). Gene networks orchestrated by MeGI: a single-factor mechanism underlying sex determination in persimmon. Plant J 98, 97-111. |
| [83] | Yang LL, Huang YT, Fu ZY, Xu QJ (2022). Research progress on the epigenetic mechanisms of sex determination in horticultural plants. Acta Hortic Sin 49, 1602-1610. (in Chinese) |
| 杨琳琳, 黄云彤, 付泽元, 徐启江 (2022). 园艺植物性别决定的表观遗传机制研究进展. 园艺学报 49, 1602-1610. | |
| [84] | Yang WL, Wang DY, Li YL, Zhang ZY, Tong SF, Li MM, Zhang X, Zhang L, Ren LW, Ma XZ, Zhou R, Sanderson BJ, Keefover-Ring K, Yin TM, Smart LB, Liu JQ, DiFazio SP, Olson M, Ma T (2021). A general model to explain repeated turnovers of sex determination in the Salicaceae. Mol Biol Evol 38, 968-980. |
| [85] | Yue JJ, Krasovec M, Kazama Y, Zhang XT, Xie WY, Zhang SC, Xu XM, Kan BL, Ming R, Filatov DA (2023). The origin and evolution of sex chromosomes, revealed by sequencing of the Silene latifolia female genome. Curr Biol 33, 2504-2514. |
| [86] | Zhang XT, Wang G, Zhang SC, Chen S, Wang YB, Wen P, Ma XK, Shi Y, Qi R, Yang Y, Liao ZY, Lin J, Lin JS, Xu XM, Chen XQ, Xu XD, Deng F, Zhao LH, Lee Yl, Wang R, Chen XY, Lin YR, Zhang JS, Tang HB, Chen J, Ming R (2020). Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell 183, 875-889. |
| [87] | Zhang XZ, Pan LS, Guo W, Li YQ, Wang WC (2022). A convergent mechanism of sex determination in dioecious plants: distinct sex-determining genes display converged regulation on floral B-class genes. Front Plant Sci 13, 953445. |
| [88] | Zhao YJ, Zhang TK, Liu CY, Huang XB, Yuan ZH (2018). Progress on sex determinant mechanism in horticultural plants. Acta Hortic Sin 45, 2228-2242. (in Chinese) |
| 赵玉洁, 张太奎, 刘翠玉, 黄贤斌, 苑兆和 (2018). 园艺植物性别决定机制研究进展. 园艺学报 45, 2228-2242. | |
| [89] | Zhou G, Chen C, Liu XH, Lu XY, Tian Y, Chen HM (2019). Research progress of sex determination in cucumber. Plant Physiol J 55, 902-914. (in Chinese) |
| 周赓, 陈宸, 刘晓虹, 卢向阳, 田云, 陈惠明 (2019). 黄瓜性别决定研究进展. 植物生理学报 55, 902-914. | |
| [90] | Zhou P, Zhang XD, Ma XY, Yue JJ, Liao ZY, Ming R (2022). Methylation related genes affect sex differentiation in dioecious and gynodioecious papaya. Hortic Res 9, uhab065. |
| [91] | Zhou R, Macaya-Sanz D, Carlson CH, Schmutz J, Jenkins JW, Kudrna D, Sharma A, Sandor L, Shu SQ, Barry K, Tuskan GA, Ma T, Liu JQ, Olson M, Smart LB, DiFazio SP (2020). A willow sex chromosome reveals convergent evolution of complex palindromic repeats. Genome Biol 21, 38. |
| [92] | Zhou YF, Minio A, Massonnet M, Solares E, Lv YD, Beridze T, Cantu? D, Gaut BS (2019). The population genetics of structural variants in grapevine domestication. Nat Plants 5, 965-979. |
| [93] | Zhu LL (2019). Analysis of MRNAs and Non-Coding RNAs Expression Involved in the Female and Male Floral Develoment of Eucommia ulmoides. Doctoral dissertation. Beijing: Chinese Academy of Forestry. pp. 1-175. (in Chinese) |
| 朱利利 (2019). 杜仲雌雄花发育相关调控基因和非编码RNA表达分析. 博士论文. 北京: 中国林业科学研究院. pp. 1-175. | |
| [94] | Zou C, Massonnet M, Minio A, Patel S, Llaca V, Karn A, Gouker F, Cadle-Davidson F, Reischg F, Fennellc A, Cantu D, Sun Q, Londo JP (2021). Multiple independent recombinations led to hermaphroditism in grapevine. Proc Natl Acad Sci USA 118, e2023548118. |
/
| 〈 |
|
〉 |