

Research Advances in Biological Functions of Plant SPL Transcription Factors
Received date: 2022-09-07
Accepted date: 2023-05-31
Online published: 2023-06-16
Squamosa promoter binding protein-like (SPL) family is a class of plant-specific transcription factors, which contain a highly conserved SBP domain consisting of two zinc finger structures and a short nuclear localization sequence. The expression of most SPL genes is regulated by microRNAs at transcription level. Based on the current research progress of SPL transcription factors, this paper summarizes the biological functions of SPLs in plant growth, development, and environmental adaptation, and discusses the future research directions of SPLs.
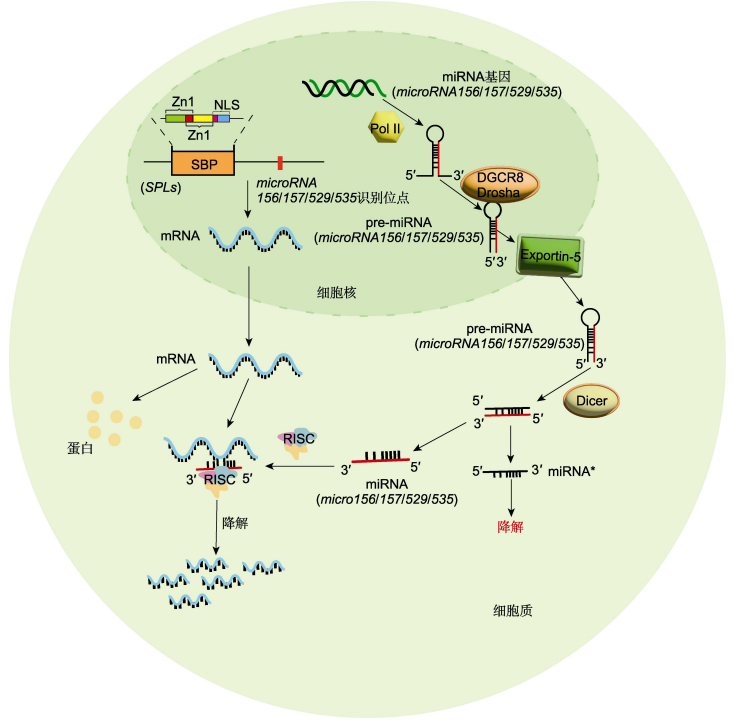
Key words: plant growth and development; SPL; transcription factor; stress responses
Xinhai Zeng , Rui Chen , Yu Shi , Chaoyue Gai , Kai Fan , Zhaowei Li . Research Advances in Biological Functions of Plant SPL Transcription Factors[J]. Chinese Bulletin of Botany, 2023 , 58(6) : 982 -997 . DOI: 10.11983/CBB22216
| [1] | Barrera-Rojas CH, Rocha GHB, Polverari L, Pinheiro Brito DA, Batista DS, Notini MM, Da Cruz ACF, Morea EGO, Sabatini S, Otoni WC, Nogueira FTS (2020). miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regenera-tion via cytokinin responses. J Exp Bot 71, 934-950. |
| [2] | Bencivenga S, Simonini S, Benková E, Colombo L (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886-2897. |
| [3] | Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, Merchant SS, Kr?mer U (2012). Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24, 738-761. |
| [4] | Birkenbihl RP, Jach G, Saedler H, Huijser P (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352, 585-596. |
| [5] | Bonnet E, He Y, Billiau K, Van De Peer Y (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566-1568. |
| [6] | Cai JJ, Liu WW, Li WQ, Zhao LJ, Chen G, Bai YY, Ma DM, Fu CX, Wang YM, Zhang XC (2022). Downregu-lation of miR156-targeted PvSPL6 in switchgrass delays flowering and increases biomass yield. Front Plant Sci 13, 834431. |
| [7] | Cao Y, Chen R, Wang WT, Wang DH, Cao XY (2021). SmSPL6 induces phenolic acid biosynthesis and affects root development in Salvia miltiorrhiza. Int J Mol Sci 22, 7895. |
| [8] | Cardon GH, H?hmann S, Nettesheim K, Saedler H, Huijser P (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12, 367-377. |
| [9] | Chao LM, Liu YQ, Chen DY, Xue XY, Mao YB, Chen XY (2017). Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol Plant 10, 735-748. |
| [10] | Chen FH, Zhang HM, Li H, Lian L, Wei YD, Lin YL, Wang LN, He W, Cai QH, Xie HG, Zhang H, Zhang JF (2023). IPA1 improves drought tolerance by activating SNAC1 in rice. BMC Plant Biol 23, 55. |
| [11] | Chen R, Cao Y, Wang WT, Li YH, Wang DH, Wang SQ, Cao XY (2021). Transcription factor SmSPL7 promotes anthocyanin accumulation and negatively regulates phenolic acid biosynthesis in Salvia miltiorrhiza. Plant Sci 310, 110993. |
| [12] | Chen WW, Jin JF, Lou HQ, Liu L, Kochian LV, Yang JL (2018). LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 248, 893-907. |
| [13] | Cheng YJ, Shang GD, Xu ZG, Yu S, Wu LY, Zhai D, Tian SL, Gao J, Wang L, Wang JW (2021). Cell division in the shoot apical meristem is a trigger for miR156 decline and vegetative phase transition in Arabidopsis. Proc Natl Acad Sci USA 118, e2115667118. |
| [14] | Cui L, Zheng FY, Wang JF, Zhang CL, Xiao FM, Ye J, Li CX, Ye ZB, Zhang JH (2020). miR156a-targeted SBP- box transcription factor SlSPL13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnol J 18, 1670-1682. |
| [15] | Dai X, Zhao PX (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39, 155-159. |
| [16] | Dong HX, Yan SL, Jing YX, Yang RZ, Zhang YW, Zhou Y, Zhu YF, Sun JQ (2021). miR156-targeted SPL9 is phosphorylated by SnRK2s and interacts with ABI5 to enhance ABA responses in Arabidopsis. Front Plant Sci 12, 708573. |
| [17] | Feng X, Zhou BJ, Wu XL, Wu HL, Zhang SL, Jiang Y, Wang YP, Zhang YQ, Cao M, Guo BS, Su SC, Hou ZX (2023). Molecular characterization of SPL gene family during flower morphogenesis and regulation in blueberry. BMC Plant Biol 23, 40. |
| [18] | Ferreirae Silva GF, Silva EM, Da Silva Azevedo M, Guivin MAC, Ramiro DA, Figueiredo CR, Carrer H, Peres LEP, Nogueira FTS (2014). MicroRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78, 604-618. |
| [19] | Feyissa BA, Amyot L, Nasrollahi V, Papadopoulos Y, Kohalmi SE, Hannoufa A (2021). Involvement of the miR156/SPL module in flooding response in Medicago sativa. Sci Rep 11, 3243. |
| [20] | Feyissa BA, Arshad M, Gruber MY, Kohalmi SE, Hannoufa A (2019). The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa. BMC Plant Biol 19, 434. |
| [21] | Gandikota M, Birkenbihl RP, H?hmann S, Cardon GH, Saedler H, Huijser P (2007). The miRNA156/157 recognition element in the 3?UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49, 683-693. |
| [22] | Garcia-Molina A, Xing SP, Huijser P (2014). Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response. BMC Plant Biol 14, 231. |
| [23] | Gou JQ, Tang CR, Chen NC, Wang H, Debnath S, Sun L, Flanagan A, Tang YH, Jiang QZ, Allen RD, Wang ZY (2019). SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol 222, 1610-1623. |
| [24] | Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512-1522. |
| [25] | Gupta A, Hua L, Zhang ZZ, Yang B, Li WL (2023). CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol J 21, 536-548. |
| [26] | Hanly A, Karagiannis J, Lu QSM, Tian LN, Hannoufa A (2020). Characterization of the role of SPL9in drought stress tolerance in Medicago sativa. Int J Mol Sci 21, 6003. |
| [27] | He YL, Fu XQ, Li L, Sun XF, Tang KX, Zhao JY (2022). AaSPL9 affects glandular trichomes initiation by positively regulating expression of AaHD1 in Artemisia annua L. Plant Sci 317, 111172. |
| [28] | Hou HM, Li J, Gao M, Singer SD, Wang H, Mao LY, Fei ZJ, Wang XP (2013). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS One 8, e59358. |
| [29] | Hu JH, Huang LY, Chen GL, Liu H, Zhang YS, Zhang R, Zhang SL, Liu JT, Hu QY, Hu FY, Wang W, Ding Y (2021a). The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice 14, 90. |
| [30] | Hu L, Chen WL, Yang W, Li XL, Zhang C, Zhang XY, Zheng L, Zhu XB, Yin JJ, Qin P, Wang YP, Ma BT, Li SG, Yuan H, Tu B (2021b). OsSPL9 regulates grain number and grain yield in rice. Front Plant Sci 12, 682018. |
| [31] | Huijser P, Klein J, L?nnig WE, Meijer H, Saedler H, Sommer H (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene SQUAMOSA in Antirrhinum majus. EMBO J 11, 1239-1249. |
| [32] | Jiang YY, Peng JR, Wang M, Su WB, Gan XQ, Jing Y, Yang XH, Lin SQ, Gao YS (2019). The role of EjSPL3, EjSPL4, EjSPL5, and EjSPL9 in regulating flowering in loquat (Eriobotrya japonica Lindl.). Int J Mol Sci 21, 248. |
| [33] | Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD, Qian Q, Li JY (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42, 541-544. |
| [34] | Jin B, Zhou XR, Jiang BL, Gu ZM, Zhang PH, Qian Q, Chen XF, Ma BJ (2015). Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice 8, 18. |
| [35] | Jones-Rhoades MW, Bartel DP, Bartel B (2006). MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57, 19-53. |
| [36] | Jung JH, Ju Y, Seo PJ, Lee JH, Park CM (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69, 577-588. |
| [37] | Lan T, Zheng YL, Su ZL, Yu SB, Song HB, Zheng XY, Lin GG, Wu WR (2019). OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3 9, 4107-4114. |
| [38] | Li HS, Ma B, Luo YW, Wei WQ, Yuan JL, Zhai CX, He NJ (2022a). The mulberry SPL gene family and the response of MnSPL7 to silkworm herbivory through activating the transcription of MnTT2L2 in the catechin biosynthesis pathway. Int J Mol Sci 23, 1141. |
| [39] | Li L, Shi F, Wang YQ, Yu XF, Zhi JJ, Guan YB, Zhao HY, Chang JL, Chen MJ, Yang GX, Wang YS, He GY (2020). TaSPL13 regulates inflorescence architecture and development in transgenic wheat (Triticum aestivum L.). Plant Sci 296, 110516. |
| [40] | Li SX, Cheng ZH, Li ZB, Dong SM, Yu XL, Zhao PJ, Liao WB, Yu X, Peng M (2022b). MeSPL9 attenuates drought resistance by regulating JA signaling and protectant metabolite contents in cassava. Theor Appl Genet 135, 817-832. |
| [41] | Li Y, He YZ, Liu ZX, Qin T, Wang L, Chen ZH, Zhang BM, Zhang HT, Li HT, Liu L, Zhang J, Yuan WY (2022c). OsSPL14 acts upstream of OsPIN1b and PILS6b to modulate axillary bud outgrowth by fine-tuning auxin transport in rice. Plant J 111, 1167-1182. |
| [42] | Lian L, Xu HB, Zhang H, He W, Cai QH, Lin YL, Wei LY, Pan LY, Xie XP, Zheng YM, Wei YD, Zhu YS, Xie HA, Zhang JF (2020). Overexpression of OsSPL14results in transcriptome and physiology changes in indica rice ‘MH86’. Plant Growth Regul 90, 265-278. |
| [43] | Liu KY, Cao J, Yu KH, Liu XY, Gao YJ, Chen Q, Zhang WJ, Peng HR, Du JK, Xin MM, Hu ZR, Guo WL, Rossi V, Ni ZF, Sun QX, Yao YY (2019). Wheat TaSPL8 modulates leaf angle through auxin and brassinosteroid signaling. Plant Physiol 181, 179-194. |
| [44] | Liu N, Tu LL, Wang LC, Hu HY, Xu J, Zhang XL (2017). MicroRNA157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol 17, 7. |
| [45] | Liu XF, Ning K, Che G, Yan SS, Han LJ, Gu R, Li Z, Weng YQ, Zhang XL (2018). CsSPL functions as an adaptor between HD-ZIP III and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J 94, 535-547. |
| [46] | Liu YT, Wu GX, Zhao YP, Wang HH, Dai ZY, Xue WC, Yang J, Wei HB, Shen RX, Wang HY (2021). DWARF53 interacts with transcription factors UB2/UB3/TSH4 to regulate maize tillering and tassel branching. Plant Physiol 187, 947-962. |
| [47] | Long JM, Liu CY, Feng MQ, Liu Y, Wu XM, Guo WW (2018). miR156-SPL modules regulate induction of somatic embryogenesis in citrus callus. J Exp Bot 69, 2979-2993. |
| [48] | Lu SF, Yang CM, Chiang VL (2011). Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J Integr Plant Biol 53, 879-891. |
| [49] | Lv ZY, Wang Y, Liu Y, Peng BW, Zhang L, Tang KX, Chen WS (2019). The SPB-box transcription factor AaSPL2 positively regulates artemisinin biosynthesis in Artemisia annua L. Front Plant Sci 10, 409. |
| [50] | Ma L, Liu XQ, Liu WH, Wen HY, Zhang YC, Pang YZ, Wang XM (2022). Characterization of Squamosa-promoter binding protein-box family genes reveals the critical role of MsSPL20 in alfalfa flowering time regulation. Front Plant Sci 12, 775690. |
| [51] | Ma Y, Xue H, Zhang F, Jiang Q, Yang S, Yue PT, Wang F, Zhang YY, Li LG, He P, Zhang ZH (2021). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol J 19, 311-323. |
| [52] | Matthews C, Arshad M, Hannoufa A (2019). Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant 165, 830-842. |
| [53] | Min XY, Luo K, Liu WX, Zhou KY, Li JY, Wei ZW (2022). Molecular characterization of the miR156/ MsSPL model in regulating the compound leaf development and abiotic stress response in alfalfa. Genes 13, 331. |
| [54] | Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42, 545-549. |
| [55] | Morea EGO, Da Silva EM, Ferreirae Silva GF, Valente GT, Barrera Rojas CH, Vincentz M, Nogueira FTS (2016). Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol 16, 40. |
| [56] | Ning K, Chen S, Huang HJ, Jiang J, Yuan HM, Li HY (2017). Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult 130, 469-481. |
| [57] | Perea-García A, Andrés-Bordería A, Huijser P, Pe?arrubia L (2021). The copper-microRNA pathway is integrated with developmental and environmental stress responses in Arabidopsis thaliana. Int J Mol Sci 22, 9547. |
| [58] | Qin MM, Zhang Y, Yang YM, Miao CB, Liu SK (2020). Seed-specific overexpression of SPL12 and IPA1 improves seed dormancy and grain size in rice. Front Plant Sci 11, 532771. |
| [59] | Rahimi A, Karami O, Balazadeh S, Offringa R (2022). miR156-independent repression of the ageing pathway by longevity-promoting AHL proteins in Arabidopsis. New Phytol 235, 2424-2438. |
| [60] | Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002). Prediction of plant microRNA targets. Cell 110, 513-520. |
| [61] | Riechmann JL, Ratcliffe OJ (2000). A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3, 423-434. |
| [62] | Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005). Specific effects of microRNAs on the plant transcriptome. Dev Cell 8, 517-527. |
| [63] | Schwechheimer C, Zourelidou M, Bevan MW (1998). Plant transcription factor studies. Annu Rev Plant Physiol Plant Mol Biol 49, 127-150. |
| [64] | Shan X, Zhang W, Huang JX, Yu FW, Qin WB, Li JB, Wang SY, Dai ZL (2021). Identification and characterization of SPL transcription factor family reveals organization and chilling-responsive patterns in cabbage (Brassica oleracea var. capitata L.). Agronomy 11, 1445. |
| [65] | Shao YL, Zhou HZ, Wu YR, Zhang H, Lin J, Jiang XY, He QJ, Zhu JS, Li Y, Yu H, Mao CZ (2019). OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 31, 1257-1275. |
| [66] | Si LZ, Chen JY, Huang XH, Gong H, Luo JH, Hou QQ, Zhou TY, Lu TT, Zhu JJ, Shangguan YY, Chen EW, Gong CX, Zhao Q, Jing YF, Zhao Y, Li Y, Cui LL, Fan DL, Lu YQ, Weng QJ, Wang YC, Zhan QL, Liu KY, Wei XH, An K, An G, Han B (2016). OsSPL13 controls grain size in cultivated rice. Nat Genet 48, 447-456. |
| [67] | Song XG, Lu ZF, Yu H, Shao GN, Xiong JS, Meng XB, Jing YH, Liu GF, Xiong GS, Duan JB, Yao XF, Liu CM, Li HQ, Wang YH, Li JY (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27, 1128-1141. |
| [68] | Stone JM, Liang XW, Nekl ER, Stiers JJ (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J 41, 744-754. |
| [69] | Sun YJ, Fu M, Wang L, Bai YX, Fang XL, Wang Q, He Y, Zeng HL (2022). OsSPLs regulate male fertility in response to different temperatures by flavonoid biosynthesis and tapetum PCD in PTGMS rice. Int J Mol Sci 23, 3744. |
| [70] | Sun YT, Wang YY, Xiao YQ, Zhang X, Du BY, Turupu M, Wang C, Yao QS, Gai SL, Huang J, Tong S, Li TH (2023). Genome-wide identification of the SQUAMOSA promoter-binding protein-like (SPL) transcription factor family in sweet cherry fruit. Int J Mol Sci 24, 2880. |
| [71] | Sunkar R, Zhu JK (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001-2019. |
| [72] | Tripathi RK, Overbeek W, Singh J (2020). Global analysis of SBP gene family in Brachypodium distachyon reveals its association with spike development. Sci Rep 10, 15032. |
| [73] | Tsuzuki M, Futagami K, Shimamura M, Inoue C, Kunimoto K, Oogami T, Tomita Y, Inoue K, Kohchi T, Yamaoka S, Araki T, Hamada T, Watanabe Y (2019). An early arising role of the microRNA156/529-SPL module in reproductive development revealed by the liverwort Marchantia polymorpha. Curr Biol 29, 3307-3314. |
| [74] | Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P (2003). SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15, 1009-1019. |
| [75] | Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41, 195-211. |
| [76] | Wang H, Wang HY (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant 8, 677-688. |
| [77] | Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin JJ, Zhu XB, Li Y, Li WT, Liu JL, Wang JC, Chen XQ, Qing H, Wang YP, Liu GF, Wang WM, Li P, Wu XJ, Zhu LH, Zhou JM, Ronald PC, Li SC, Li JY, Chen XW (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026-1028. |
| [78] | Wang JW, Czech B, Weigel D (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738-749. |
| [79] | Wang JW, Ye YJ, Xu M, Feng LG, Xu LA (2019). Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol 19, 370. |
| [80] | Wang L, Ming LC, Liao KY, Xia CJ, Sun SY, Chang Y, Wang HK, Fu DB, Xu CH, Wang ZJ, Li X, Xie WB, Ouyang YD, Zhang QL, Li XH, Zhang QH, Xiao JH, Zhang QF (2021a). Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol Plant 14, 1168-1184. |
| [81] | Wang L, Sun SY, Jin JY, Fu DB, Yang XF, Weng XY, Xu CG, Li XH, Xiao JH, Zhang QF (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci USA 112, 15504-15509. |
| [82] | Wang M, Mo ZH, Lin RZ, Zhu CC (2021b). Characterization and expression analysis of the SPL gene family during floral development and abiotic stress in pecan (Carya illinoinensis). Peer J 9, e12490. |
| [83] | Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, Zhang GQ, Fu XD (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44, 950-954. |
| [84] | Wang SL, Ren XX, Xue JQ, Xue YQ, Cheng XD, Hou XG, Zhang XX (2020a). Molecular characterization and expression analysis of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE gene family in Paeonia suffruticosa. Plant Cell Rep 39, 1425-1441. |
| [85] | Wang YM, Liu WW, Wang XW, Yang RJ, Wu ZY, Wang H, Wang L, Hu ZB, Guo SY, Zhang HL, Lin JX, Fu CX (2020b). miR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic Res 7, 118. |
| [86] | Wei BY, Zhang JZ, Pang CX, Yu H, Guo DS, Jiang H, Ding MX, Chen ZY, Tao Q, Gu HY, Qu LJ, Qin GJ (2015). The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res 25, 121-134. |
| [87] | Wu G, Poethig RS (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539-3547. |
| [88] | Wu ZY, Cao YP, Yang RJ, Qi TX, Hang YQ, Lin H, Zhou GK, Wang ZY, Fu CX (2016). Switchgrass SBP-box transcription factors PvSPL1 and 2 function redundantly to initiate side tillers and affect biomass yield of energy crop. Biotechnol Biofuels 9, 101. |
| [89] | Xing SP, Salinas M, H?hmann S, Berndtgen R, Huijser P (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935-3950. |
| [90] | Xu ML, Hu TQ, Zhao JF, Park MY, Earley KW, Wu G, Yang L, Poethig RS (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12, e1006263. |
| [91] | Xu YJ, Xu HX, Wall MM, Yang JZ (2020). Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 112, 2734-2747. |
| [92] | Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17, 268-278. |
| [93] | Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009). SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347-361. |
| [94] | Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol 337, 49-63. |
| [95] | Yang RJ, Liu WW, Sun Y, Sun ZC, Wu ZY, Wang YM, Wang MQ, Wang HL, Bai SQ, Fu CX (2022). LATERAL BRANCHING OXIDOREDUCTASE, one novel target gene of Squamosa promoter binding protein-like 2, regulates tillering in switchgrass. New Phytol 235, 563-575. |
| [96] | Yao SZ, Yang ZR, Yang RX, Huang Y, Guo G, Kong XY, Lan Y, Zhou T, Wang H, Wang WM, Cao XF, Wu JG, Li Y (2019a). Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol Plant 12, 1114-1122. |
| [97] | Yao T, Park BS, Mao HZ, Seo JS, Ohama N, Li Y, Yu N, Mustafa NFB, Huang CH, Chua NH (2019b). Regulation of flowering time by SPL10/MED25 module in Arabidopsis. New Phytol 224, 493-504. |
| [98] | Yin HB, Hong GJ, Li LY, Zhang XY, Kong YZ, Sun ZT, Li JM, Chen JP, He YQ (2019). miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 109, 632-642. |
| [99] | Yu ZX, Wang LJ, Zhao B, Shan CM, Zhang YH, Chen DF, Chen XY (2015). Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol Plant 8, 98-110. |
| [100] | Yuan H, Qin P, Hu L, Zhan SJ, Wang SF, Gao P, Li J, Jin MY, Xu ZY, Gao Q, Du AP, Tu B, Chen WL, Ma BT, Wang YP, Li SG (2019). OsSPL18 controls grain weight and grain number in rice. J Genet Genomics 46, 41-51. |
| [101] | Yue EK, Li C, Li Y, Liu Z, Xu JH (2017). miR529a modulates panicle architecture through regulating SQUAMOSA PROMOTER BINDING-LIKE genes in rice (Oryza sativa). Plant Mol Biol 94, 469-480. |
| [102] | Yun JX, Sun ZX, Jiang Q, Wang YN, Wang C, Luo YQ, Zhang FR, Li X (2022). The miR156b-GmSPL9d module modulates nodulation by targeting multiple core nodulation genes in soybean. New Phytol 233, 1881-1899. |
| [103] | Zhang LL, Huang YY, Zheng YP, Liu XX, Zhou SX, Yang XM, Liu SL, Li Y, Li JL, Zhao SL, Wang H, Ji YP, Zhang JW, Pu M, Zhao ZX, Fan J, Wang WM (2022). Osa- miR535 targets SQUAMOSA promoter binding protein-like 4 to regulate blast disease resistance in rice. Plant J 110, 166-178. |
| [104] | Zhang XF, Yang CY, Lin HX, Wang JW, Xue HW (2021). Rice SPL12 coevolved with GW5 to determine grain shape. Sci Bull 66, 2353-2357. |
| [105] | Zhang Y, Schwarz S, Saedler H, Huijser P (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63, 429-439. |
| [106] | Zhao BB, Xu MY, Zhao YP, Li YY, Xu H, Li CY, Kong DX, Xie YR, Zheng ZG, Wang BB, Wang HY (2022a). Over-expression of ZmSPL12 confers enhanced lodging resistance through transcriptional regulation of D1 in maize. Plant Biotechnol J 20, 622-624. |
| [107] | Zhao JL, Shi M, Yu J, Guo CK (2022b). SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol 22, 59. |
| [108] | Zhou MQ, Tang W (2019). MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol Genet Genomics 294, 379-393. |
/
| 〈 |
|
〉 |