

Research Progress in Sirtuin Protein Family in Plants
Received date: 2022-11-02
Accepted date: 2023-05-31
Online published: 2023-05-31
The sirtuin protein family of plants is a class of enzymes that depend on β-nicotinamide adenine dinucleotide (β-NAD+) to catalyze the removal of Nε-acetyl and Nε-acyl (acetyl, crotonyl, butylyl and 2-hydroxy-isobutylyl) from non-histone and histone lysine, respectively, and has the function of regulating the activity of non-histone protein and gene expression. Members of the sirtuin protein family have been shown to play important epigentic regulatory roles in the growth and development of rice (Oryza sativa), soybean (Glycine max) and tomato (Solanum lycopersicum), and in response to salt, cold, heat and pathogen stresses. Here we summarized the recent research advances in the enzyme activity, catalytic substrates, cell localization and function of plant sirtuins, which will serve as a reference for understanding the epigenetic regulation mechanism and studying the new function of sirtuin.
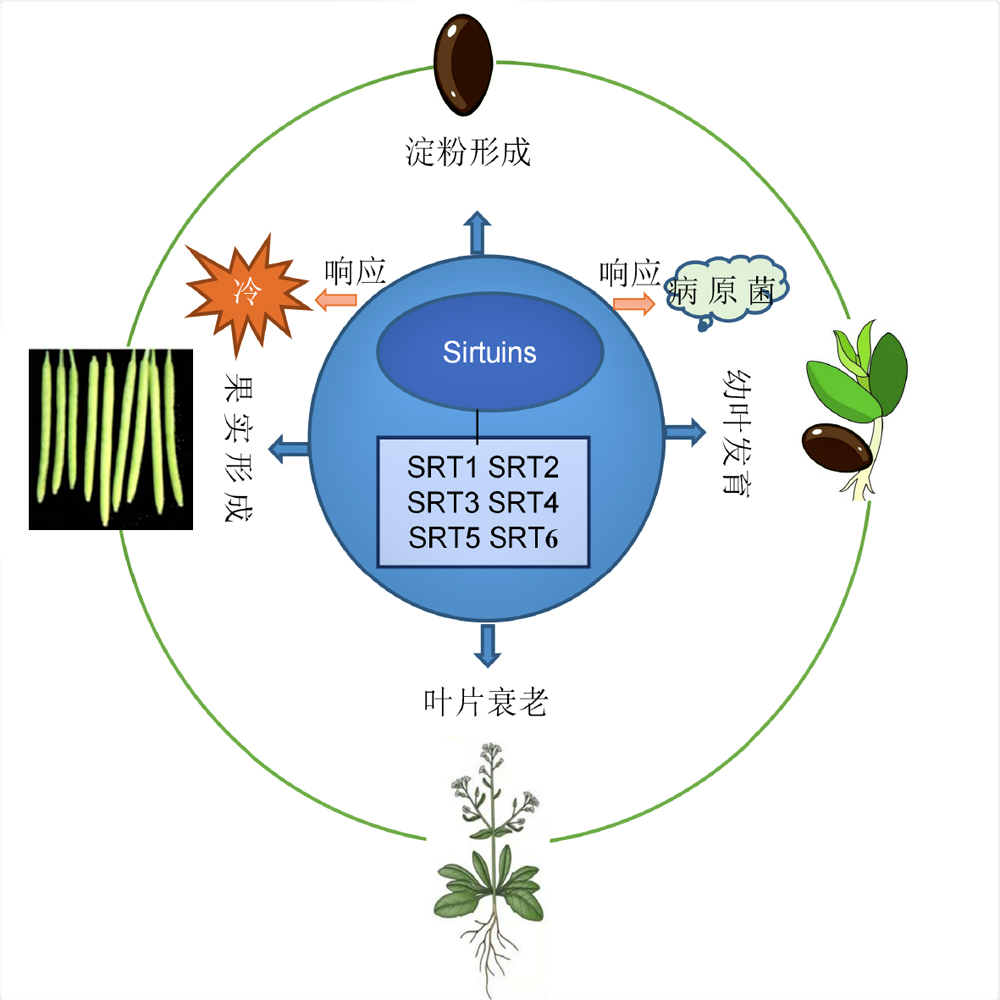
Key words: plant; sirtuins; deacylation; epigenetic regulation; development; stress
Lufang Su , Ping Wang , Shun Li , Yan Cai , Dandan Guo , Qin Liu , Xiaoyun Liu . Research Progress in Sirtuin Protein Family in Plants[J]. Chinese Bulletin of Botany, 2023 , 58(6) : 998 -1007 . DOI: 10.11983/CBB22255
| [1] | 陈聪, 宋江波, 孟刚, 童晓玲, 代方银, 鲁成 (2014). 家蚕sirtuin家族基因的鉴定及系统发生与表达芯片分析. 中国农业科学 47, 2659-2670. |
| [2] | 陈威, 杨颖增, 陈锋, 周文冠, 舒凯 (2019). 表观遗传修饰介导的植物胁迫记忆. 植物学报 54, 779-785. |
| [3] | Bheda P, Jing H, Wolberger C, Lin HN (2016). The substrate specificity of sirtuins. Annu Rev Biochem 85, 405-429. |
| [4] | Blander G, Guarente L (2004). The Sir2 family of protein deacetylases. Annu Rev Biochem 73, 417-435. |
| [5] | Bourque S, Dutartre A, Hammoudi V, Blanc S, Dahan J, Jeandroz S, Pichereaux C, Rossignol M, Wendehenne D (2011). Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol 192, 127-139. |
| [6] | Bruscalupi G, Di Micco P, Failla CM, Pascarella G, Morea V, Saliola M, De Paolis A, Venditti S, Mauro ML (2023). Arabidopsis thaliana sirtuins control proliferation and glutamate dehydrogenase activity. Plant Physiol Biochem 194, 236-245. |
| [7] | Cantó C, Auwerx J (2011). NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb Symp Quant Biol 76, 291-298. |
| [8] | Carabetta VJ, Cristea IM (2017). Regulation, function, and detection of protein acetylation in bacteria. J Bacteriol 199, e00107-17. |
| [9] | Carrico C, Meyer JG, He WJ, Gibson BW, Verdin E (2018). The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab 27, 497-512. |
| [10] | Chen B, Zang WW, Wang J, Huang YJ, He YH, Yan LL, Liu JJ, Zheng WP (2015). The chemical biology of sirtuins. Chem Soc Rev 44, 5246-5264. |
| [11] | Chen XY, Xu QT, Duan YH, Liu H, Chen XL, Huang JB, Luo CX, Zhou DX, Zheng L (2021). Ustilaginoidea virens modulates lysine 2-hydroxyisobutyrylation in rice flowers during infection. J Integr Plant Biol 63, 1801-1814. |
| [12] | Chen Z, Luo L, Chen RF, Hu HH, Pan YF, Jiang HB, Wan X, Jin H, Gong YM (2018). Acetylome profiling reveals extensive lysine acetylation of the fatty acid metabolism pathway in the diatom Phaeodactylum tricornutum. Mol Cell Proteomics 17, 399-412. |
| [13] | Chu JS, Chen Z (2018). Molecular identification of histone acetyltransferases and deacetylases in lower plant Marchantia polymorpha. Plant Physiol Biochem 132, 612-622. |
| [14] | Chung PJ, Kim YS, Park SH, Nahm BH, Kim JK (2009). Subcellular localization of rice histone deacetylases in organelles. FEBS Let 583, 2249-2254. |
| [15] | Cucurachi M, Busconi M, Morreale G, Zanetti A, Bavaresco L, Fogher C (2012). Characterization and differential expression analysis of complete coding sequences of Vitis vinifera L. sirtuin genes. Plant Physiol Biochem 54, 123-132. |
| [16] | De Block M, Van Lijsebettens M (2011). Energy efficiency and energy homeostasis as genetic and epigenetic components of plant performance and crop productivity. Curr Opin Plant Biol 14, 275-282. |
| [17] | De Block M, Verduyn C, De Brouwer D, Cornelissen M (2005). Poly (ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J 41, 95-106. |
| [18] | Dutta A, Abmayr SM, Workman JL (2016). Diverse activities of histone acylations connect metabolism to chromatin function. Mol Cell 63, 547-552. |
| [19] | Fan W, Luo JY (2010). SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell 39, 247-258. |
| [20] | Fang CY, Zhang H, Wan J, Wu YY, Li K, Jin C, Chen W, Wang SC, Wang WS, Zhang HW, Zhang P, Zhang F, Qu LH, Liu XQ, Zhou DX, Luo J (2016). Control of leaf senescence by an MeOH-jasmonates cascade that is epigenetically regulated by OsSRT1 in rice. Mol Plant 9, 1366-1378. |
| [21] | Fang Y, Tang S, Li XL (2019). Sirtuins in metabolic and epigenetic regulation of stem cells. Trends Endocrinol Metabol 30, 177-188. |
| [22] | Feldman JL, Dittenhafer-Reed KE, Denu JM (2012). Sirtuin catalysis and regulation. J Biol Chem 287, 42419-42427. |
| [23] | Greiss S, Gartner A (2009). Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells 28, 407-415. |
| [24] | Guarente L (2011). The logic linking protein acetylation and metabolism. Cell Metab 14, 151-153. |
| [25] | Hashida SN, Takahashi H, Uchimiya H (2009). The role of NAD biosynthesis in plant development and stress responses. Ann Bot 103, 819-824. |
| [26] | Huang LM, Sun QW, Qin FJ, Li C, Zhao Y, Zhou DX (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144, 1508-1519. |
| [27] | Imai SI, Armstrong CM, Kaeberlein M, Guarente L (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795-800. |
| [28] | Kaelin WG Jr, McKnight SL (2013). Influence of metabolism on epigenetics and disease. Cell 153, 56-69. |
| [29] | Kang M, Abdelmageed H, Lee S, Reichert A, Mysore KS, Allen RD (2013). AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J 76, 481-493. |
| [30] | K?nig AC, Hartl M, Pham PA, Laxa M, Boersema PJ, Orwat A, Kalitventseva I, Pl?chinger M, Braun HP, Leister D, Mann M, Wachter A, Fernie AR, Finkemeier I (2014). The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol 164, 1401-1414. |
| [31] | Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T (2008). A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 105, 3374-3379. |
| [32] | Lee K, Park OS, Jung SJ, Seo PJ (2016). Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J Plant Physiol 191, 95-100. |
| [33] | Li SC, Zheng WP (2018). Mammalian sirtuins SIRT4 and SIRT7. Prog Mol Biol Transl Sci 154, 147-168. |
| [34] | Liu XY, Wei W, Zhu WJ, Su LF, Xiong ZY, Zhou M, Zheng Y, Zhou DX (2017). Histone deacetylase AtSRT1 links metabolic flux and stress response in Arabidopsis. Mol Plant 10, 1510-1522. |
| [35] | Lu Y, Xu QT, Liu Y, Yu Y, Cheng ZY, Zhao Y, Zhou DX (2018). Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol 19, 144. |
| [36] | Narita T, Weinert BT, Choudhary C (2019). Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol 20, 156-174. |
| [37] | O'Callaghan C, Vassilopoulos A (2017). Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 16, 1208-1218. |
| [38] | Rajabi N, Galleano I, Madsen AS, Olsen CA (2018). Targeting sirtuins: substrate specificity and inhibitor design. Prog Mol Biol Transl Sci 154, 25-69. |
| [39] | Reed SM, Quelle DE (2015). p53 acetylation: regulation and consequences. Cancers 7, 30-69. |
| [40] | Ringel AE, Tucker SA, Haigis MC (2018). Chemical and physiological features of mitochondrial acylation. Mol Cell 72, 610-624. |
| [41] | Sridha S, Wu KQ (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46, 124-133. |
| [42] | Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP (2011). The deacetylase SIRT1 promotes membrane localization and activation of AKT and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal 4, ra46. |
| [43] | Tang WS, Zhong L, Ding QQ, Dou YN, Li WW, Xu ZS, Zhou YB, Chen J, Chen M, Ma YZ (2022). Histone deacetylase AtSRT2 regulates salt tolerance during seed germination via repression of vesicle-associated membrane protein 714 (VAMP714) in Arabidopsis. New Phytol 234, 1278-1293. |
| [44] | Vall-Llaura N, Torres R, Lindo-García V, Mu?oz P, Munné-Bosch S, Larrigaudière C, Teixidó N, Giné- Bordonaba J (2021). PbSRT1 and PbSRT2 regulate pear growth and ripening yet displaying a species-specific regulation in comparison to other Rosaceae spp. Plant Sci 308, 110925. |
| [45] | Wang CZ, Gao F, Wu JG, Dai JL, Wei H, Li Y (2010). Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51, 1291-1299. |
| [46] | Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, Irani K (2010). SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res 38, 832-845. |
| [47] | Yang C, Shen WJ, Chen HF, Chu LT, Xu YC, Zhou XC, Liu CL, Chen CM, Zeng JH, Liu J, Li QF, Gao CJ, Charron JB, Luo M (2018). Characterization and subcellular localization of histone deacetylases and their roles in response to abiotic stresses in soybean. BMC Plant Biol 18, 226. |
| [48] | Zhang F, Wang LK, Ko EE, Shao K, Qiao H (2018). Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30, 153-166. |
| [49] | Zhang H, Lu Y, Zhao Y, Zhou DX (2016). OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci 248, 28-36. |
| [50] | Zhang H, Zhao Y, Zhou DX (2017). Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res 45, 12241-12255. |
| [51] | Zhao LM, Lu JX, Zhang JX, Wu PY, Yang SG, Wu KQ (2015). Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front Plant Sci 5, 760. |
| [52] | Zhao S, Zhang XR, Li HT (2018). Beyond histone acetylation-writing and erasing histone acylations. Curr Opin Struct Biol 53, 169-177. |
| [53] | Zhao SM, Xu W, Jiang WQ, Yu W, Lin Y, Zhang TF, Yao J, Zhou L, Zeng YX, Li H, Li YX, Shi J, An WL, Hancock SM, He FC, Qin LX, Chin J, Yang PY, Chen X, Lei QY, Xiong Y, Guan KL (2010). Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000-1004. |
| [54] | Zheng WP (2020). Review: the plant sirtuins. Plant Sci 293, 110434. |
| [55] | Zhong XC, Zhang H, Zhao Y, Sun QW, Hu YF, Peng H, Zhou DX (2013). The rice NAD+-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS One 8, e66807. |
/
| 〈 |
|
〉 |