

Double-stranded Labelled Oligo-FISH in Rice Chromosomes
Received date: 2022-03-24
Accepted date: 2022-08-04
Online published: 2022-08-19
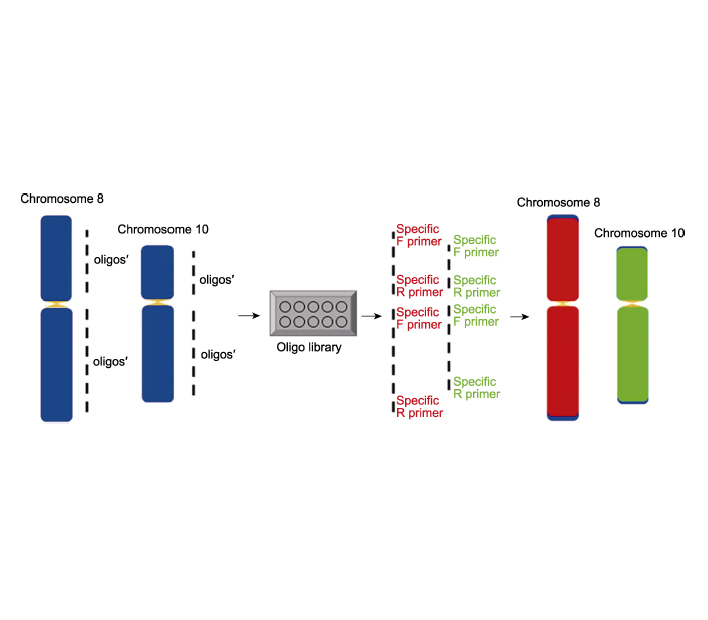
The technique of identification and preparation of chromosome(s) are important tools in genetic research. Oligonucleotide fluorescence in situ hybridization (oligo-FISH) is an emerging chromosome identification technique in recent years. Flexible and efficient probes are the key factors in the process of oligo-FISH. The labelling process of traditional single-stranded oligo probes (ss-oligos) is complicated and the cost of obtaining individual probes is high. By improving the ss-oligo probes labelling process, we obtain the probes by PCR amplification with specific primers targeting the whole chromosome (fragment), which simplifies the probe labelling process, reduces the cost and improves the labelling efficiency. In this study, we describe in detail the synthesis and labelling of a modified double-stranded labelled oligo probes (ds-oligos) library in rice (Oryza sativa), the preparation of mitotic chromosomes and the hybridization process of ds-oligo probes. By designing gradient experiments, the optimal denaturation time and temperature of chromosome and oligo probe in rice were found to be 85°C for 3.5 min and 90°C for 6 min, respectively. This is the first study to establish a chromosomal double-stranded labelled oligo-FISH system in rice, which provides a powerful tool for the preparation and precise identification of chromosomes in a variety of plants.
Key words: rice; oligo-FISH; mitotic
Shang Sun , Yingying Hu , Yangshuo Han , Chao Xue , Zhiyun Gong . Double-stranded Labelled Oligo-FISH in Rice Chromosomes[J]. Chinese Bulletin of Botany, 2023 , 58(3) : 433 -439 . DOI: 10.11983/CBB22055
| [1] | Anderson LK, Stack SM, Mitchell JB (1982). An investiga-tion of the basis of a current hypothesis for the lack of G- banding in plant chromosomes. Exp Cell Res 138, 433-436. |
| [2] | Ban C, Chung S, Park DS, Shim YB (2004). Detection of protein-DNA interaction with a DNA probe: distinction between single-strand and double-strand DNA-protein interaction. Nucleic Acids Res 32, e110. |
| [3] | Bi YF, Zhao QZ, Yan WK, Li MX, Liu YX, Cheng CY, Zhang L, Yu XQ, Li J, Qian CT, Wu YF, Chen JF, Lou QF (2020). Flexible chromosome painting based on multiplex PCR of oligo-nucleotides and its application for comparative chro-mosome analyses in Cucumis. Plant J 102, 178-186. |
| [4] | Braz GT, do Vale Martins L, Zhang T, Albert PS, Birchler JA, Jiang JM, (2020). A universal chromosome identifica-tion system for maize and wild Zea species. Chromosome Res 28, 183-194. |
| [5] | Braz GT, He L, Zhao HN, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018). Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal Karyo-typic and chromosomal evolution. Genetics 208, 513-523. |
| [6] | do Vale Martins L, Yu F, Zhao HN, Dennison T, Lauter N, Wang HY, Deng ZH, Thompson A, Semrau K, Rouillard JM, Birchler JA, Jiang JM (2019). Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10, 4604. |
| [7] | Fuchs J, Houben A, Brandes A, Schubert I (1996). Chro-mosome ‘painting’ in plants—a feasible technique? Chromosoma 104, 315-320. |
| [8] | Hou LL, Xu M, Zhang T, Xu ZH, Wang WY, Zhang JX, Yu MM, Ji W, Zhu CW, Gong ZY, Gu MH, Jiang JM, Yu HX (2018). Chromosome painting and its applications in culti-vated and wild rice. BMC Plant Biol 18, 110. |
| [9] | Huang W, Du Y, Zhao X, Jin WW (2016). B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.). BMC Plant Biol 16, 88. |
| [10] | Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995). Metaphase and interphase fluorescence in situ hybridiza-tion mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92, 4487-4491. |
| [11] | Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res 27, 153-165. |
| [12] | Jiang JM, Gill BS (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057-1068. |
| [13] | Landegent JE, Jansen in de Wal N, Dirks RW, van der Ploeg M (1987). Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet 77, 366-370. |
| [14] | Langer-Safer PR, Levine M, Ward DC (1982). Immunological method for mapping genes on drosophila polytene chromosomes. Proc Natl Acad Sci USA 79, 4381-4385. |
| [15] | Li ZA, Bi YF, Wang X, Wang YZ, Yang SQ, Zhang ZT, Chen JF, Lou QF (2018). Chromosome identification in Cucumis anguria revealed by cross-species single-copy gene FISH. Genome 61, 397-404. |
| [16] | Liu XY, Sun S, Wu Y, Zhou Y, Gu SW, Yu HX, Yi CD, Gu MH, Jiang JM, Liu B, Zhang T, Gong ZY (2020). Dual- color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101, 112-121. |
| [17] | Lou QF, Zhang YX, He YH, Li J, Jia L, Cheng CY, Guan W, Yang SQ, Chen JF (2014). Single-copy gene-based chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. Plant J 78, 169-179. |
| [18] | Mukai Y, Nakahara Y, Yamamoto M (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total ge-nomic and highly repeated DNA probes. Genome 36, 489-494. |
| [19] | Ouyang S, Zhu W, Hamilton J, Lin HN, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35, D883-D887. |
| [20] | ?imonikova D, Něme?ková A, ?í?ková J, Brown A, Swennen R, Dole?el J, H?ibová E (2020). Chromosome painting in cultivated bananas and their wild relatives (Musa spp.) reveals differences in chromosome structure. Int J Mol Sci 21, 7915. |
| [21] | Tang XM, Bao WD, Zhang WL, Cheng ZK (2007). Identifica-tion of chromosomes from multiple rice genomes using a universal molecular cytogenetic marker system. J Integr Plant Biol 49, 953-960. |
| [22] | Trent JM, Thompson FH (1987). Methods for chromosome banding of human and experimental tumors in vitro. Methods Enzymol 151, 267-279. |
| [23] | Xin HY, Zhang T, Wu YF, Zhang WL, Zhang PD, Xi ML, Jiang JM (2020). An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J 101, 253-264. |
| [24] | Zhang T, Liu GQ, Zhao HN, Braz GT, Jiang JM (2021). Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J 19, 1967-1978. |
| [25] | Zhao QZ, Meng Y, Wang PQ, Qin XD, Cheng CY, Zhou JG, Yu XQ, Li J, Lou QF, Jahn M, Chen JF (2021). Reconst-ruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J 107, 1243-1259. |
/
| 〈 |
|
〉 |