

QTL Mapping of Candidate Genes for Heading Date in Rice
Received date: 2022-05-31
Accepted date: 2022-07-25
Online published: 2022-07-25
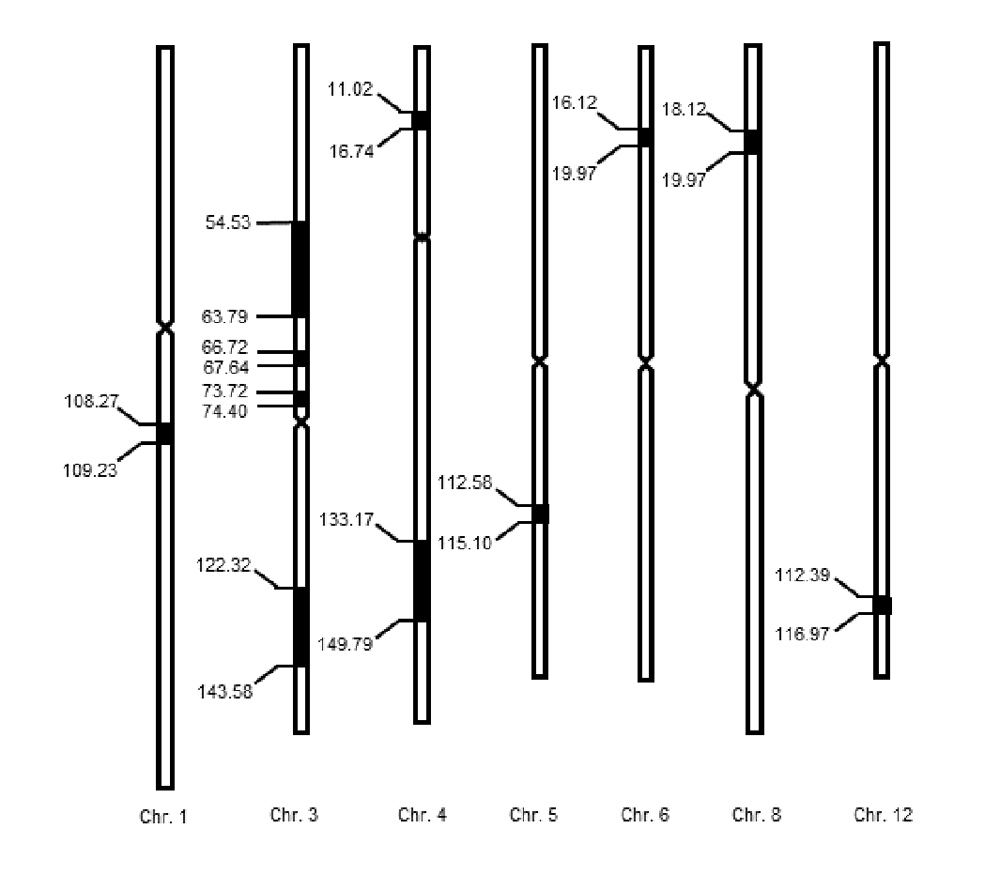
The heading date is an important trait that determines the yield and quality of rice, and plays an important role in the process of rice breeding, seed production, introduction and domestication. In this study, F1 was obtained by a cross between Nekken2 (Oryza sativa subsp. japonica cv. ‘Nekken2’) and Huazhan (O. sativa subsp. indica cv. ‘HZ’), and 120 recombination inbred lines (RILs) were obtained through successive multi-generational selfing. The population of RILs was used as experimental material. Under the condition of conventional water and fertilizer management, the time of heading stage of 120 lines was analyzed. Using the high-density genetic map constructed by this population, the QTLs for the heading date was mapped and analyzed. A total of 11 QTLs were detected on chromosomes 1, 3, 4, 5, 6, 8, and 12, respectively, and one of the LOD values was as high as 5.75. By analyzing the candidate genes in the QTLs interval, the related genes that may affect the heading date were screened out, and real-time quantitative PCR was used for gene expression analysis. The expression levels of six genes, LOC_Os03g03070, LOC_Os03g50310, LOC_Os03g55389, LOC_Os04g55510, LOC_Os08g07740, and LOC_Os08g01670 significantly different between parents, and the expression of LOC_Os03g50310 in Nekken2 was 3.6 times higher than that in HZ. In addition, the sequencing analysis showed that there were 4 differences in the 5'UTR, CDS region and 3'UTR of the candidate gene LOC_Os03g50310 between the parents, of which the single nucleotide polymorphisms (SNP) difference in the CDS region caused a single amino acid change. This study provides new clues for further cloning of heading date-related genes and cultivar selection by mining the QTL loci related to heading date in rice.
Wei Heping , Lu Tao , Jia Qiwei , Deng Fei , Zhu Hao , Qi Zehua , Wang Yuxi , Ye Hanfei , Yin Wenjing , Fang Yuan , Mu Dan , Rao Yuchun . QTL Mapping of Candidate Genes for Heading Date in Rice[J]. Chinese Bulletin of Botany, 2022 , 57(5) : 588 -595 . DOI: 10.11983/CBB22114
| [1] | 戴高兴 (2012). 超级杂交稻协优9308重组自交系抽穗期QTL定位及其与产量性状关系的研究. 博士论文. 北京: 中国农业科学院. pp. 1-56. |
| [2] | 蒋丹, 洪广成, 陈倩, 刘石锋, 秦小健 (2019). 水稻抽穗期分子调控研究进展. 分子植物育种 17, 7071-7077. |
| [3] | 孙志超 (2018). 11个微效水稻抽穗期QTL的检测与验证. 硕士论文. 北京: 中国农业科学院. pp. 21-25. |
| [4] | 杨德卫, 陈壬杰, 程朝平, 郑向华, 叶宁, 叶新福, 黄凤凰 (2019). 水稻抽穗期基因的鉴定与遗传调控网络研究与分析. 分子植物育种 17, 4656-4660. |
| [5] | 杨锟 (2019). 水稻穗发芽和抽穗期的QTL定位. 硕士论文. 南京: 南京农业大学. pp. 34-36. |
| [6] | 张立成, 李懿星, 王天抗, 邱牡丹, 宋书锋, 董皓, 李磊, 刘建丰, 李莉 (2020). 水稻抽穗期基因OsDof6功能的初步研究. 中国水稻科学 34, 397-405. |
| [7] | Adachi S, Yoshikawa K, Yamanouchi U, Tanabata T, Sun J, Ookawa T, Yamamoto T, Sage RF, Hirasawa T, Yonemaru J (2017). Fine mapping of Carbon Assimilation Rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front Plant Sci 8, 60. |
| [8] | Bian XF, Liu X, Zhao ZG, Jiang L, Gao H, Zhang YH, Zheng M, Chen LM, Liu SJ, Zhai HQ, Wan JM (2011). Heading date gene, dth3 controlled late flowering in O. glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep 30, 2243-2254. |
| [9] | Dai C, Xue HW (2010). Rice early flowering 1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signaling. EMBO J 29, 1916-1927. |
| [10] | Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18, 926-936. |
| [11] | Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M (2013). Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J 76, 36-46. |
| [12] | Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43, 1096-1105. |
| [13] | Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang ZY, Li JJ, Li ZC, Paek NC (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6, 1877-1888. |
| [14] | Kwon CT, Yoo SC, Koo BH, Cho SH, Park JW, Zhang ZY, Li JJ, Li ZC, Paek NC (2014). Natural variation in Early flowering 1 contributes to early flowering in japonica rice under long days. Plant Cell Environ 37, 101-112. |
| [15] | Liu H, Dong SY, Sun DY, Liu W, Gu FW, Liu YZ, Guo T, Wang H, Wang JF, Chen ZQ (2016). CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS One 11, e0166249. |
| [16] | Liu TM, Liu HY, Zhang H, Xing XZ (2013). Validation and characterization of Ghd7.1, a major quantitative trait locus with pleiotropic effects on spikelets per panicle, plant height, and heading date in rice (Oryza sativa L.). J Integr Plant Biol 55, 917-927. |
| [17] | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods 25, 402-408. |
| [18] | Matsoukas IG, Massiah AJ, Thomas B (2012). Florigenic and antiflorigenic signaling in plants. Plant Cell Physiol 53, 1827-1842. |
| [19] | Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M (2011). Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66, 603-612. |
| [20] | McCouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H (1997). Report on QTL nomenclature. Rice Genet Newsl 14, 11-13. |
| [21] | Monna L, Lin H, Kojima S, Sasaki T, Yano M (2002). Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor Appl Genet 104, 772-778. |
| [22] | Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008). Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105, 3646-3651. |
| [23] | Takahashi Y, Shomura A, Sasaki T, Yano M (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc Natl Acad Sci USA 98, 7922-7927. |
| [24] | Tsuji H, Tachibana C, Tamaki S, Taoka KI, Kyozuka J, Shimamoto K (2015). Hd3a promotes lateral branching in rice. Plant J 82, 256-266. |
| [25] | Wei XJ, Xu JF, Guo HN, Jiang L, Chen SH, Yu CY, Zhou ZL, Hu PS, Zhai HQ, Wan JM (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153, 1747-1758. |
| [26] | Wu WX, Zheng XM, Lu GW, Zhong ZZ, Gao H, Chen LP, Wu CY, Wang HJ, Wang Q, Zhou KN, Wang JL, Wu FQ, Zhang X, Guo XP, Cheng ZJ, Lei CL, Lin QB, Jiang L, Wang HY, Ge S, Wan JM (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110, 2775-2780. |
| [27] | Xue WY, Xing Y, Weng X, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40, 761-767. |
| [28] | Yamamoto T, Lin HX, Sasaki T, Yano M (2000). Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics 154, 885-891. |
| [29] | Yang Y, Fu DB, Zhu CM, He YZ, Zhang HJ, Liu T, Li XH, Wu CY (2015). The RING-finger ubiquitin ligase HAF1 mediates Heading date 1 degradation during photoperiodic flowering in rice. Plant Cell 27, 2455-2468. |
| [30] | Yano M, Kojima S, Takahashi Y, Lin HX, Sasaki T (2001). Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127, 1425-1429. |
| [31] | Zhang J, Zhou XC, Yan WH, Zhang ZY, Lu L, Han ZM, Zhao H, Liu HY, Song P, Hu Y, Shen GJ, He Q, Guo SB, Gao GQ, Wang GW, Xing YZ (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol 208, 1056-1066. |
| [32] | Zong WB, Ren D, Huang MH, Sun KL, Feng JL, Zhao J, Xiao DD, Xie WH, Liu SQ, Zhang H, Qiu R, Tang WJ, Yang RQ, Chen HY, Xie XR, Chen LT, Liu YG, Guo JX (2021). Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol 229, 1635-1649. |
/
| 〈 |
|
〉 |