

Alternative Splicing of Plant Genes: Full of Change, Sail with Wind
Received date: 2022-03-07
Accepted date: 2022-05-10
Online published: 2022-05-10
Alternative splicing is a process in which the mRNA precursor produces a variety of mature transcripts through different splicing mode. It is an important way in post transcriptional regulation in eukaryotic cells. Plant genes also can produce diverse transcripts via alternative splicing on the same gene, so as to regulate a variety of biological processes. In plants, the research on this field lags behind that in animals, but great progresses have been made in recent years. In this review, we summarized the biological significance, splicing patterns, research methods of the alternative splicing, as well as its roles in the regulation in the growth, development and responses to environmental stresses. Some perspectives and prospects are give for this research field.
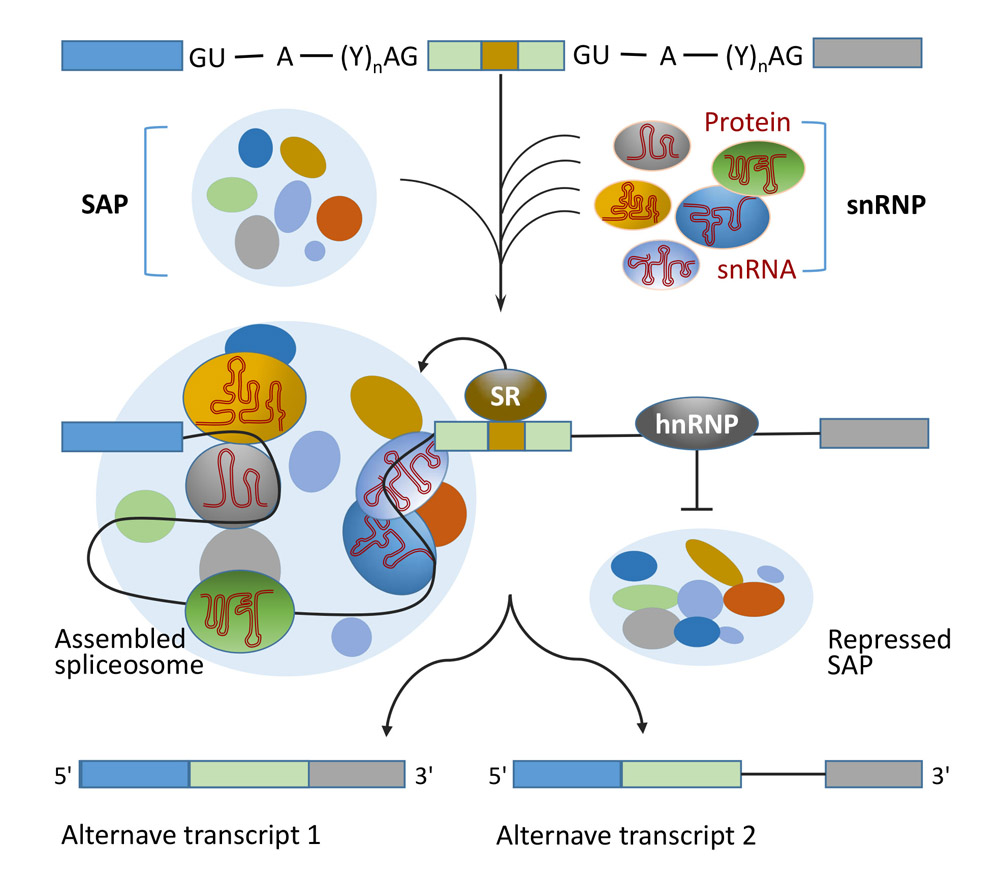
Key words: plants; gene; alternative splicing; transcripts; function
Hao Xuefeng , Wu Zhangjing , Ma Tian , Jin Zhuping , Pei Yanxi . Alternative Splicing of Plant Genes: Full of Change, Sail with Wind[J]. Chinese Bulletin of Botany, 2022 , 57(5) : 661 -672 . DOI: 10.11983/CBB22041
| [1] | 蔡芳芳, 邵长生, 孙玉强 (2022). 可变剪切在植物成花转换中的作用. 植物学报 57, 69-79. |
| [2] | Abascal F, Ezkurdia I, Rodriguez-Rivas J, Rodriguez JM, del Pozo A, Vázquez J, Valencia A, Tress ML (2015). Alternatively spliced homologous exons have ancient origins and are highly expressed at the protein level. PLoS Comput Biol 11, e1004325. |
| [3] | Barta A, Marquez Y, Brown JWS (2012). Challenges in plant alternative splicing. In: Stamm S, Smith CWJ, Lührmann R, eds. Alternative Pre-mRNA Splicing: Theory and Protocols. Weinheim: Wiley-VCH Verlag. pp. 79-89. |
| [4] | Barta A, Sommergruber K, Thompson D, Hartmuth K, Matzke MA, Matzke AJM (1986). The expression of a nopaline synthase-human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol Biol 6, 347-357. |
| [5] | Blencowe BJ (2006). Alternative splicing: new insights from global analyses. Cell 126, 37-47. |
| [6] | Carvalho RF, Carvalho SD, Duque P (2010). The plant- specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol 154, 772-783. |
| [7] | Carvalho RF, Szakonyi D, Simpson CG, Barbosa ICR, Brown JWS, Baena-González E, Duque P (2016). The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 28, 1910-1925. |
| [8] | Carvalho SD, Saraiva R, Maia TM, Abreu IA, Duque P (2012). XBAT35, a novel Arabidopsis RING E3 ligase exhibiting dual targeting of its splice isoforms, is involved in ethylene-mediated regulation of apical hook curvature. Mol Plant 5, 1295-1309. |
| [9] | Chamala S, Feng GQ, Chavarro C, Barbazuk WB (2015). Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Frontiers Bioeng Biotech 3, 33. |
| [10] | Chaudhary S, Jabre I, Reddy ASN, Staiger D, Syed NH (2019). Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci 24, 496-506. |
| [11] | Chen T, Cui P, Chen H, Ali S, Zhang SD, Xiong LM (2013). A KH-domain RNA-binding protein interacts with FIERY2/ CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 9, e1003875. |
| [12] | Chen WJ, Moore MJ (2015). Spliceosomes. Curr Biol 25, R181-R183. |
| [13] | Daszkowska-Golec A, Skubacz A, Marzec M, Slota M, Kurowska M, Gajecka M, Gajewska P, P?ociniczak T, Sitko K, Pacak A, Szweykowska-Kulinska Z, Szarejko I (2017). Mutation in HvCBP20 (cap binding protein 20) adapts barley to drought stress at phenotypic and transcriptomic levels. Front Plant Sci 8, 942. |
| [14] | Day IS, Golovkin M, Palusa SG, Link A, Ali GS, Thomas J, Richardson DN, Reddy ASN (2012). Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: insights into regulated splicing. Plant J 71, 936-947. |
| [15] | de la Fuente van Bentem S, Vossen JH, Vermeer JEM, de Vroomen MJ, Gadella TWJ Jr, Haring MA, Cornelissen BJC (2003). The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol 133, 702-712. |
| [16] | Deng X, Gu LF, Liu CY, Lu TC, Lu FL, Lu ZK, Cui P, Pei YX, Wang BC, Hu SN, Cao XF (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci USA 107, 19114-19119. |
| [17] | Dikaya V, El Arbi N, Rojas-Murcia N, Nardeli SM, Goretti D, Schmid M (2021). Insights into the role of alternative splicing in plant temperature response. J Exp Bot 72, 7384-7403. |
| [18] | Duque P (2011). A role for SR proteins in plant stress responses. Plant Signal Behav 6, 49-54. |
| [19] | Early P, Rogers J, Davisa M, Calame K, Bond M, Wall R, Hood L (1980). Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell 20, 313-319. |
| [20] | Eckardt NA (2013). The plant cell reviews alternative splicing. Plant Cell 25, 3639. |
| [21] | Emami S, Arumainayagam D, Korf I, Rose AB (2013). The effects of a stimulating intron on the expression of heterologous genes in Arabidopsis thaliana. Plant Biotechnol J 11, 555-563. |
| [22] | Fouquet R, Martin F, Fajardo DS, Gault CM, Gómez E, Tseung CW, Policht T, Hueros G, Settles AM (2011). Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell 23, 4280-4297. |
| [23] | Hasegawa M, Miura T, Kuzuya K, Inoue A, Won Ki S, Horinouchi S, Yoshida T, Kunoh T, Koseki K, Mino K, Sasaki R, Yoshida M, Mizukami T (2011). Identification of SAP155 as the target of GEX1A (herboxidiene), an antitumor natural product. ACS Chem Biol 6, 229-233. |
| [24] | Heinrich B, Zhang ZY, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S (2009). Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem 284, 14303-14315. |
| [25] | Huang CF, Miki D, Tang K, Zhou HR, Zheng ZM, Chen W, Ma ZY, Yang L, Zhang H, Liu RY, He XJ, Zhu JK (2013). A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet 9, e100-3779. |
| [26] | Hugouvieux V, Kwak JM, Schroeder JI (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477-487. |
| [27] | Iida K, Shionyu M, Suso Y (2008). Alternative splicing at NAGNAG acceptor sites shares common properties in land plants and mammals. Mol Biol Evol 25, 709-718. |
| [28] | Izaurralde E, Lewis J, Mcguigan C, Jankowska M, Darzynkiewicz E, Mattaj IW (1994). A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78, 657-668. |
| [29] | Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, Dinh HQ, Barta A, Brown JWS (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40, 2454-2469. |
| [30] | Kim YO, Pan SO, Jung CH, Kang H (2007). A zinc finger-containing glycine-rich RNA binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol 48, 1170-1181. |
| [31] | Koncz C, deJong F, Villacorta N, Szakonyi D, Koncz Z (2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front Plant Sci 3, 9. |
| [32] | Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y (2010). Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336-339. |
| [33] | Kong XX, Ma L, Yang LM, Chen Q, Xiang N, Yang YP, Hu XY (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins Arabidopsis CBP20 and CBP80 modulate the salt stress response. J Proteome Res 13, 2495-2510. |
| [34] | Laloum T, Martín G, Duque P (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci 23, 140-150. |
| [35] | Lazar G, Goodman HM (2000). The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol Biol 42, 571-581. |
| [36] | Lee BH, Kapoor A, Zhu JH, Zhu JK (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre- mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18, 1736-1749. |
| [37] | Lin JY, Zhu ZQ (2021). Plant responses to high temperature: a view from pre-mRNA alternative splicing. Plant Mol Biol 105, 575-583. |
| [38] | Ling Y, Alshareef S, Butt H, Lozano-Juste J, Li LX, Galal AA, Moustafa A, Momin AA, Tashkandi M, Richardson DN, Fujii H, Arold S, Rodriguez PL, Duque P, Mahfouz MM (2017). Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J 89, 291-309. |
| [39] | Ling Y, Serrano N, Gao G, Atia M, Mokhtar M, Woo YH, Bazin J, Veluchamy A, Benhamed M, Crespi M, Gehring C, Reddy ASN, Mahfouz MM (2018). Thermopriming triggers splicing memory in Arabidopsis. J Exp Bot 69, 2659-2675. |
| [40] | Lopato S, Mayeda A, Krainer AR, Barta A (1996). Pre- mRNA splicing in plants: characterization of Ser/Arg splicing factors. Proc Natl Acad Sci USA 93, 3074-3079. |
| [41] | Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22, 1184-1195. |
| [42] | Mei WB, Boatwright L, Feng GQ, Schnable JC, Barbazuk WB (2017a). Evolutionarily conserved alternative splicing across monocots. Genetics 207, 465-480. |
| [43] | Mei WB, Liu SZ, Schnable JC, Yeh CT, Springer NM, Schnable PS, Barbazuk WB (2017b). A comprehensive analysis of alternative splicing in paleopolyploid maize. Front Plant Sci 8, 694. |
| [44] | Merkin J, Russell C, Chen P, Burge CB (2012). Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593-1599. |
| [45] | Mulligan GJ, Guo W, Wormsley S, Helfman DM (1992). Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem 267, 25480-25487. |
| [46] | Nieto Moreno N, Giono LE, Cambindo Botto AE, Mu?oz MJ, Kornblihtt AR (2015). Chromatin, DNA structure and alternative splicing. FEBS Lett 589, 3370-3378. |
| [47] | Nilsen TW, Graveley BR (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457-463. |
| [48] | Pajoro A, Severing E, Angenent GC, Immink RGH (2017). Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol 18, 102. |
| [49] | Palusa SG, Ali GS, Reddy ASN (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49, 1091-1107. |
| [50] | Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ (2005). Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21, 73-77. |
| [51] | Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068-1071. |
| [52] | Pei YX, Chen ZJ, Cao JS, Chen XJ, Liu XH (2004). Cytoplasmic male sterility of tuber mustard is associated with the alternative spliced mitochondrial T gene transcripts. Chin Sci Bull 49, 2481-2486. |
| [53] | Pei YX, Qiao ZJ, Chen XJ, Chen ZJ, Cao JS, Yu X, Ma LH, Liu XH (2008). T1243, an alternative transcript of the mitochondrial T gene in Brassica juncea var. tumida, causes pollen abortion in Arabidopsis thaliana. Plant Sci 175, 793-798. |
| [54] | Pelechano V, Wei W, Steinmetz LM (2013). Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497, 127-131. |
| [55] | Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24, 4930-4947. |
| [56] | Punzo P, Grillo S, Batelli G (2020). Alternative splicing in plant abiotic stress responses. Biochem Soc Trans 48, 2117-2126. |
| [57] | Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JWS, Jarmolowski A (2010). Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res 38, 265-278. |
| [58] | Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010). ABA perception and signaling. Trends Plant Sci 15, 395-401. |
| [59] | Reddy ASN, Marquez Y, Kalyna M, Barta A (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657-3683. |
| [60] | Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A, Lumbreras V, González-Guzmán M, Antoni R, Rodriguez PL, Baena- González E (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase 1 signaling in Arabidopsis. Plant Cell 25, 3871-3884. |
| [61] | Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL (2000). Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671-684. |
| [62] | Singer GA, Wu JJ, Yan P, Plass C, Huang TH, Davuluri RV (2008). Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genomics 9, 349. |
| [63] | Song QA, Catlin NS, Brad Barbazuk W, Li S (2019). Computational analysis of alternative splicing in plant genomes. Gene 685, 186-195. |
| [64] | Srikar C, Feng G, Carolina C, Brad BW (2015). Genome- wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front Bioeng Biotech 3, 33. |
| [65] | Staudt AC, Wenkel S (2011). Regulation of protein function by ‘microProteins’. EMBO Rep 12, 35-42. |
| [66] | Swiezewski S, Liu F, Magusin A, Dean C (2009). Cold- induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799-802. |
| [67] | Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S (2007). Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol 48, 1036-1049. |
| [68] | Tharun S (2009). Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol 272, 149-189. |
| [69] | Wachter A, Rühl C, Stauffer E (2012). The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front Plant Sci 3, 81. |
| [70] | Wan RX, Bai R, Yan CY, Lei JL, Shi YG (2019). Structures of the catalytically activated yeast spliceosome reveal the mechanism of branching. Cell 177, 339-351. |
| [71] | Wang BB, Brendel V (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol 5, R102. |
| [72] | Wang PC, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang HM, Tao WA, Zhu JK (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110, 11205-11210. |
| [73] | Wang YY, Xiong F, Ren QP, Wang XL (2020). Regulation of flowering transition by alternative splicing: the role of the U2 auxiliary factor. J Exp Bot 71, 751-758. |
| [74] | Wang ZJ, Ji HT, Yuan BJ, Wang SF, Su C, Yao B, Zhao HT, Li X (2015). ABA signaling is fine-tuned by antagonistic HAB1 variants. Nat Commun 6, 8138. |
| [75] | Werneke JM, Chatfield JM, Ogren WL (1989). Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1, 815-825. |
| [76] | Wu XH, Liu M, Downie B, Liang C, Ji GL, Li QQ, Hunt AG (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA 108, 12533-12538. |
| [77] | Xing DH, Wang YJ, Hamilton M, Ben-Hur A, Reddy ASN (2015). Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27, 3294-3308. |
| [78] | Xiong F, Ren JJ, Yu Q, Wang YY, Lu CC, Kong LJ, Otegui MS, Wang XL (2019). AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5and FLC in Arabidopsis. New Phytol 223, 277-292. |
| [79] | Xiong LM, Gong ZZ, Rock CD, Subramanian S, Guo Y, Xu WY, Galbraith D, Zhu JK (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1, 771-781. |
| [80] | Yang XZ, Zhang HY, Li L (2012). Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J 70, 421-431. |
| [81] | Yu HP, Tian CH, Yu Y, Jiao YL (2016). Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol Plant 9, 749-752. |
| [82] | Zhang GJ, Guo GW, Hu XD, Zhang Y, Li QY, Li RQ, Zhuang RH, Lu ZK, He ZQ, Fang XD, Chen L, Tian W, Tao Y, Kristiansen K, Zhang XQ, Li SG, Yang HM, Wang J, Wang J (2010). Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20, 646-654. |
| [83] | Zhang RX, Calixto CPG, Marquez Y, Venhuizen P, Tzioutziou NA, Guo WB, Spensley M, Entizne JC, Lewandowska D, Ten Have S, Frei Dit Frey N, Hirt H, James AB, Nimmo HG, Barta A, Kalyna M, Brown JWS (2017). A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res 45, 5061-5073. |
| [84] | Zhang WT, Du BJ, Liu D, Qi XT (2014). Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem Biophys Res Commun 455, 312-317. |
| [85] | Zhang ZL, Zhang SP, Zhang Y, Wang X, Li D, Li QL, Yue MH, Li Q, Zhang YE, Xu YY, Xue YB, Chong K, Bao SL (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396-411. |
/
| 〈 |
|
〉 |