

A Review on the Mechanism of Ribosome Stress Response in Plants
Received date: 2021-07-23
Accepted date: 2021-10-12
Online published: 2021-10-12
The nucleolus is a key structure in the nucleus of eukaryotic cells. Ribosome biogenesis initially takes place in the nucleolus, which involves a series of complex reactions and requires the participation of many nucleolus-associated factors. Abnormal ribosome biogenesis usually leads to disordered nucleolus structure, cell cycle arrest, cell senescence, and even cell death. The mechanism of ribosomal stress response has been studied well in mammals, while it remains largely unknown in plant cells. Nevertheless, it has been found that members of the plant-specific NAC transcription factor family are involved in various intracellular stress responses, including ribosomal stress, in plant cells. In addition, we previously reported a coordination mechanism between auxin system and ribosome biogenesis for modulating plant development. In this review, we discussed potential mechanisms of ribosome stress response in plant cells through combining the knowledge of ribosomal stress response pathways in mammals.
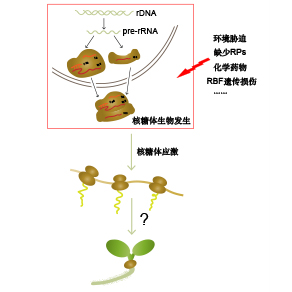
Key words: ribosome; ribosomal stress; NAC transcription factor; MIDASIN 1; auxin
Jingwen Wang , Xingjun Wang , Changle Ma , Pengcheng Li . A Review on the Mechanism of Ribosome Stress Response in Plants[J]. Chinese Bulletin of Botany, 2022 , 57(1) : 80 -89 . DOI: 10.11983/CBB21120
| [1] | Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J 64, 960-976. |
| [2] | Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Söding J, Westhof E, Wilson DN, Beckmann R (2010). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci USA 107, 19748-19753. |
| [3] | Balazadeh S, Wu AH, Mueller-Roeber B (2010). Salt- triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behav 5, 733-735. |
| [4] | Barzilai A, Yamamoto KI (2004). DNA damage responses to oxidative stress. DNA Repair 3, 1109-1115. |
| [5] | Baßler J, Hurt E (2019). Eukaryotic ribosome assembly. Annu Rev Biochem 88, 281-306. |
| [6] | Baßler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010). The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38, 712-721. |
| [7] | Ben-Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524-1529. |
| [8] | Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574-585. |
| [9] | Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010). The nucleolus under stress. Mol Cell 40, 216-227. |
| [10] | Byrne ME (2009). A role for the ribosome in development. Trends Plant Sci 14, 512-519. |
| [11] | Cao J, Geballe AP (1996). Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol 16, 603-608. |
| [12] | Cech TT (2000). The ribosome is a ribozyme. Science 289, 878-879. |
| [13] | Cerezo E, Plisson-Chastang C, Henras AK, Lebaron S, Gleizes PE, O'Donohue MF, Romeo Y, Henry Y (2019). Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip Rev RNA 10, e1516. |
| [14] | Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS (2011). Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol 31, 4007-4021. |
| [15] | Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007). Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029-5037. |
| [16] | Clowes FAL (2000). Pattern in root meristem development in angiosperms. New Phytol 146, 83-94. |
| [17] | Costanzo M, Nishikawa JL, Tang XJ, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004). CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899-913. |
| [18] | Eichler DC, Craig N (1994). Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49, 197-239. |
| [19] | Ernst HA, Olsen AN, Skriver K, Larsen S, Lo Leggio L (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5, 297-303. |
| [20] | Esposito D, Crescenzi E, Sagar V, Loreni F, Russo A, Russo G (2014). Human rpL3 plays a crucial role in cell response to nucleolar stress induced by 5-FU and L-OHP. Oncotarget 5, 11737-11751. |
| [21] | Garapati P, Xue GP, Munné-Bosch S, Balazadeh S (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol 168, 1122-1139. |
| [22] | Gómez-Herreros F, Rodríguez-Galán O, Morillo-Huesca M, Maya D, Arista-Romero M, de la Cruz J, Chávez S, Muñoz-Centeno MC (2013). Balanced production of ribosome components is required for proper G1/S transition in Saccharomyces cerevisiae. J Biol Chem 288, 31689-31700. |
| [23] | Grandi P, Rybin V, Baßler J, Petfalski E, Strauß D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E (2002). 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10, 105-115. |
| [24] | Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6, 225-242. |
| [25] | Holmberg Olausson K, Nistér M, Lindström MS (2012). p53-dependent and -independent nucleolar stress re- sponses. Cells 1, 774-798. |
| [26] | Horn HF, Vousden KH (2007). Coping with stress: multiple ways to activate p53. Oncogene 26, 1306-1316. |
| [27] | Hu WW, Feng ZH, Levine AJ (2012). The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer 3, 199-208. |
| [28] | James A, Wang YB, Raje H, Rosby R, DiMario P (2014). Nucleolar stress with and without p53. Nucleus 5, 402-426. |
| [29] | Klinge S, Woolford JL Jr (2019). Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20, 116-131. |
| [30] | Kornprobst M, Turk M, Kellner N, Cheng JD, Flemming D, Koš-Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R, Hurt E (2016). Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380-393. |
| [31] | Koš M, Tollervey D (2010). Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37, 809-820. |
| [32] | Kressler D, Hurt E, Bergler H, Baßler J (2012). The power of AAA-ATPases on the road of pre-60S ribosome maturation-molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823, 92-100. |
| [33] | Kubbutat MHG, Jones SN, Vousden KH (1997). Regulation of p53 stability by Mdm2. Nature 387, 299-303. |
| [34] | Lafontaine DLJ (2015). Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22, 11-19. |
| [35] | Law GL, Raney A, Heusner C, Morris DR (2001). Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276, 38036-38043. |
| [36] | Lee MM, Schiefelbein J (1999). WEREWOLF, a MYB- related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473-483. |
| [37] | Li K, Zhou XM, Sun XP, Li GH, Hou L, Zhao SZ, Zhao CZ, Ma CL, Li PC, Wang XJ (2021). Coordination between MIDASIN 1-mediated ribosome biogenesis and auxin modulates plant development. J Exp Bot 72, 2501-2513. |
| [38] | Li PC, Li K, Wang J, Zhao CZ, Zhao SZ, Hou L, Xia H, Ma CL, Wang XJ (2019). The AAA-ATPase MIDASIN 1 functions in ribosome biogenesis and is essential for embryo and root development. Plant Physiol 180, 289-304. |
| [39] | Lu L, Yi HM, Chen C, Yan SC, Yao H, He GC, Li GF, Jiang YQ, Deng T, Deng XY (2018). Nucleolar stress: is there a reverse version? J Cancer 9, 3723-3727. |
| [40] | Maekawa S, Ishida T, Yanagisawa S (2018). Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30, 209-227. |
| [41] | Mills EW, Green R (2017). Ribosomopathies: there's strength in numbers. Science 358, eaan2755. |
| [42] | Ohbayashi I, Lin CY, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M (2017). Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29, 2644-2660. |
| [43] | Olson MOJ (2004). Sensing cellular stress: another new function for the nucleolus? Sci STKE 224, pe10. |
| [44] | Palm D, Streit D, Shanmugam T, Weis BL, Ruprecht M, Simm S, Schleiff E (2019). Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res 47, 1880-1895. |
| [45] | Pelletier J, Thomas G, Volarević S (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18, 51-63. |
| [46] | Petricka JJ, Nelson TM (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol 144, 173-186. |
| [47] | Rosado A, Sohn EJ, Drakakaki G, Pan SQ, Swidergal A, Xiong YQ, Kang BH, Bressan RA, Raikhel NV (2010). Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 22, 143-158. |
| [48] | Sáez-Vásquez J, Delseny M (2019). Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31, 1945-1967. |
| [49] | Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73, 39-85. |
| [50] | Shiloh Y (2001). ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 11, 71-77. |
| [51] | Shinohara N, Ohbayashi I, Sugiyama M (2014). Involvement of rRNA biosynthesis in the regulation of CUC1 gene expression and pre-meristematic cell mound formation during shoot regeneration. Front Plant Sci 5, 159. |
| [52] | Szakonyi D, Byrne ME (2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65, 269-281. |
| [53] | Takahashi H, Takahashi A, Naito S, Onouchi H (2012). BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28, 2231-2241. |
| [54] | Tomecki R, Sikorski PJ, Zakrzewska-Placzek M (2017). Comparison of preribosomal RNA processing pathways in yeast, plant and human cells-focus on coordinated action of endo- and exoribonucleases. FEBS Lett 591, 1801-1850. |
| [55] | Unfried I, Gruendler P (1990). Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18, 4011. |
| [56] | Unfried I, Stocker U, Gruendler P (1989). Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Res 17, 7513. |
| [57] | Wang WJ, Ryu KH, Bruex A, Barron C, Schiefelbein J (2020). Molecular basis for a cell fate switch in response to impaired ribosome biogenesis in the Arabidopsis root epidermis. Plant Cell 32, 2402-2423. |
| [58] | Weis BL, Kovacevic J, Missbach S, Schleiff E (2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci 20, 729-740. |
| [59] | Weis BL, Palm D, Missbach S, Bohnsack MT, Schleiff E (2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21, 415-425. |
| [60] | Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009). Suppressor of gamma response 1 ( SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106, 12843-12848. |
| [61] | Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M (2013). ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 14, 817-822. |
| [62] | Yusupova G, Yusupov M (2014). High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem 83, 467-486. |
| [63] | Zemp I, Kutay U (2007). Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581, 2783-2793. |
/
| 〈 |
|
〉 |