

Research Progress in Seed Development, Dormancy and Germination Regulated by Cytokinin
Received date: 2020-08-03
Accepted date: 2021-01-05
Online published: 2021-01-22
Seed germination is an important stage in the establishment, growth and propagation of plants, and plays a critical role in the life cycle of seed plants. Seed dormancy is formed during development, and reaches its peak at physiological maturity. The phytohormone regulation of seed dormancy and germination may be a highly conserved mechanism in seed plants. Cytokinin (CK) is one of the most important signal molecules in plants, and regulates many aspects of plant growth and development. The bioactive CK levels are controlled by a balance among biosynthesis, activation, deactivation, re-activation and degradation, and seed development, dormancy and germination are regulated by bioactive CK levels and signaling pathways. Here, we mainly summarize the research progresses of CK biosynthesis and catabolism, signaling, and regulation on seed development, dormancy and germination. In addition, we also propose some scientific questions that need further addressed in this field to provide some information for understanding the molecular mechanism of seed development, dormancy and germination regulated by CK.
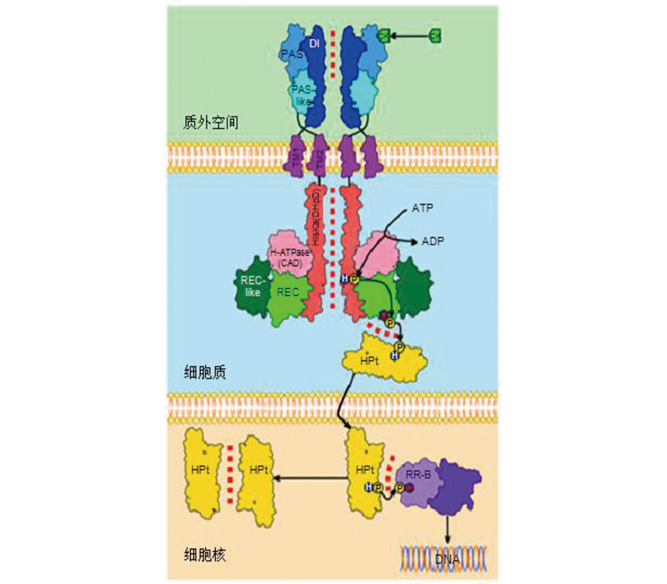
Key words: cytokinin; metabolism; molecular mechanism; seed dormancy and germination; signaling
Songquan Song , Jun Liu , Hua Yang , Wenhu Zhang , Qi Zhang , Jiadong Gao . Research Progress in Seed Development, Dormancy and Germination Regulated by Cytokinin[J]. Chinese Bulletin of Botany, 2021 , 56(2) : 218 -231 . DOI: 10.11983/CBB20141
| [1] | 邓志军, 宋松泉 (2008). ABA对黑黄檀种子萌发的抑制作用以及其他植物激素对ABA的拮抗作用. 云南植物研究 30,440-446. |
| [2] | 邓志军, 宋松泉, 艾训儒, 姚兰 (2019). 植物种子保存和检测的原理与技术. 北京: 科学出版社. pp.22-74. |
| [3] | 宋松泉, 刘军, 黄荟, 伍贤进, 徐恒恒, 张琪, 李秀梅, 梁娟 (2020a). 赤霉素代谢与信号转导及其调控种子萌发与休眠的分子机制. 中国科学: 生命科学 50,599-615. |
| [4] | 宋松泉, 刘军, 徐恒恒, 刘旭, 黄荟 (2020b). 脱落酸代谢与信号传递及其调控种子休眠与萌发的分子机制. 中国农业科学 53,857-873. |
| [5] | 徐恒恒, 黎妮, 刘树君, 王伟青, 王伟平, 张红, 程红焱, 宋松泉 (2014). 种子萌发及其调控的研究进展. 作物学报 40,1141-1156. |
| [6] | Araújo S, Pagano A, Dondi D, Lazzaroni S, Pinela E, Macovei A, Balestrazzi A (2019). Metabolic signatures of germination triggered by kinetin in Medicago truncatula. Sci Rep 9, 10466. |
| [7] | Arkhipov DV, Lomin SN, Myakushina YA, Savelieva EM, Osolodkin DI, Romanov GA (2019). Modeling of protein-protein interactions in cytokinin signal transduction. Int J Mol Sci 20,2096. |
| [8] | Baskin CC, Baskin JM (2014). Seeds:Ecology, Biogeography, and Evolution of Dormancy and Germination, 2nd edn. Amsterdam: Academic Press. pp.5-77. |
| [9] | Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110,E435- E444. |
| [10] | Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H (2013). Physiology of Development,Germination and Dormancy, 3rd edn. New York: Springer. pp.27-83. |
| [11] | Cairns JRK, Esen A (2010). β-glucosidases. Cell Mol Life Sci 67,3389-3405. |
| [12] | Chen L, Zhao JQ, Song JC, Jameson PE (2020). Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol J 18,614-630. |
| [13] | Chitnis VR, Gao F, Yao Z, Jordan MC, Park S, Ayele BT (2014). After-ripening induced transcriptional changes of hormonal genes in wheat seeds: the cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS One 9,e87543. |
| [14] | Corbineau F, Xia Q, Bailly C, EI-Maarouf-Bouteau H (2014). Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5,539. |
| [15] | Day RC, Herridge RP, Ambrose BA, Macknight RC (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148,1964-1984. |
| [16] | Deng Y, Dong HL, Mu JY, Ren B, Zheng BL, Ji ZD, Yang WC, Liang Y, Zuo JR (2010). Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22,1232-1248. |
| [17] | Eastwood D, Tavener RJA, Laidman DL (1969). Sequential action of cytokinin and gibberellic acid in wheat aleurone tissue. Nature 221,1267. |
| [18] | Feng J, Wang C, Chen QG, Chen H, Ren B, Li XM, Zuo JR (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4,1529. |
| [19] | Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P (2011). Evolution of cytokinin biosynthesis and degradation. J Exp Bot 62,2431-2452. |
| [20] | Guan CM, Wang XC, Feng J, Hong SL, Liang Y, Ren B, Zuo JR (2014). Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of ABSCISIC ACID INSENSITIVE 5 protein in Arabidopsis. Plant Physiol 164, 1515-1526. |
| [21] | Hallmark HT, Rashotte AM (2019). Review—cytokinin response factors: responding to more than cytokinin. Plant Sci 289,110251. |
| [22] | Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59,75-83. |
| [23] | Ho?ek P, Hoyerová K, Kiran NS, Dobrev PI, Zahajská L, Filepová R, Motyka V, Müller K, Kamínek M (2020). Distinct metabolism of N-glucosides of isopentenyladenine and trans-zeatin determines cytokinin metabolic spectrum in Arabidopsis. New Phytol 225,2423-2438. |
| [24] | Hothorn M, Dabi T, Chory J (2011). Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol 7,766-768. |
| [25] | Hou BK, Lim EK, Higgins GS, Bowles DJ (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279,47822-47832. |
| [26] | Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18,3073-3087. |
| [27] | Hwang I, Sheen J, Müller B (2012). Cytokinin signaling networks. Annu Rev Plant Biol 63,353-380. |
| [28] | Jameson PE, Dhandapani P, Novak O, Song JC (2016). Cytokinins and expression of SWEET, SUT, CWINV and AAP genes increase as pea seeds germinate. Int J Mol Sci 17,2013. |
| [29] | Jameson PE, Song JC (2016). Cytokinin: a key driver of seed yield. J Exp Bot 67,593-606. |
| [30] | Kakimoto T (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol 42,677-685. |
| [31] | Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem 279,14049-14054. |
| [32] | Keshishian EA, Rashotte AM (2015). Plant cytokinin signaling. Essays Biochem 58,13-27. |
| [33] | Khan AA (1968). Inhibition of gibberellic acid-induced germination by abscisic acid and reversal by cytokinins. Plant Physiol 43,1463-1465. |
| [34] | Kieber JJ, Schaller GE (2014). Cytokinins. Arabidopsis Book 12,e0168. |
| [35] | Kieber JJ, Schaller GE (2018). Cytokinin signaling in plant development. Development 145,dev149344. |
| [36] | Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445,652-655. |
| [37] | Li YJ, Cheng HY, Song SQ (2009). Effects of temperature, after-ripening, stratification, and scarification plus hormone treatments on dormancy release and germination of Acer truncatum seeds. Seed Sci Technol 37,554-562. |
| [38] | Liu HX, Stone SL (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22,2630-2641. |
| [39] | Liu XD, Zhang H, Zhao Y, Feng ZY, Li Q, Yang HQ, Luan S, Li JM, He ZH (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110,15485-15490. |
| [40] | Liu ZN, Yuan L, Song XY, Yu XL, Sundaresan V (2017). AHP2, AHP3, and AHP5 act downstream of CKI1 in Arabidopsis female gametophyte development. J Exp Bot 68,3365-3373. |
| [41] | Lomin SN, Krivosheev DM, Steklov MY, Arkhipov DV, Osolodkin DI, Schmülling T, Romanov GA (2015). Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J Exp Bot 66,1851-1863. |
| [42] | Lomin SN, Myakushina YA, Kolachevskaya OO, Getman IA, Arkhipov DV, Savelieva EM, Osolodkin DI, Romanov GA (2018). Cytokinin perception in potato: new features of canonical players. J Exp Bot 69,3839-3853. |
| [43] | Lur HS, Setter TL (1993). Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103,273- 280. |
| [44] | M?h?nen AP, Higuchi M, Tormakangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T (2006). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16, 1116-1122. |
| [45] | Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Gronlund AL, Wanchoo- Kohli A, Thomas SG, Bennett MJ, Lohmann JU, Blázquez MA, Alabadí D (2015). Genome wide binding site analysis reveals transcriptional coactivation of cytokinin- responsive genes by DELLA proteins. PLoS Genet 11,e1005337. |
| [46] | Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37,128-138. |
| [47] | Mok MC, Martin RC, Dobrev PI, Vanková R, Ho PS, Yonekura-Sakakibara K, Sakakibara H, Mok DWS (2005). Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition. Plant Physiol 137,1057-1066. |
| [48] | Müller B, Sheen J (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453,1094-1097. |
| [49] | Nguyen HN, Perry L, Kisiala A, Olechowski H, Emery RJN (2020). Cytokinin activity during early kernel development corresponds positively with yield potential and later stage ABA accumulation in field-grown wheat ( Triticum aestivum L.). Planta 252,76. |
| [50] | Nonogaki H (2014). Seed dormancy and germination- emerging mechanisms and new hypotheses. Front Plant Sci 5,233. |
| [51] | Nonogaki H (2017). Seed biology updates—highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8,524. |
| [52] | Nonogaki H (2019). Seed germination and dormancy: the classic story, new puzzles, and evolution. J Integr Plant Biol 61,541-563. |
| [53] | Pekárová B, Szmitkowska A, Dopitová R, Degtjarik O, ?ídek L, Hejátko J (2016). Structural aspects of multistep phosphorelay-mediated signaling in plants. Mol Plant 9,71-85. |
| [54] | Pekarova B, Szmitkowska A, Houser J, Wimmerova M, Hejátko J (2018). Cytokinin and ethylene signaling. In: Hejátko J, Hakoshima T, eds. Plant Structural Biology:Hormonal Regulations. Cham: Springer. pp.165-200. |
| [55] | Ren B, Liang Y, Deng Y, Chen QG, Zhang J, Yang XH, Zuo JR (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res 19, 1178- 1190. |
| [56] | Rijavec T, Dermastia M (2010). Cytokinins and their function in developing seeds. Acta Chim Slov 57,617-629. |
| [57] | Romanov GA, Lomin SN, Schmülling T (2018). Cytokinin signaling: from the ER or from the PM? That is the question! New Phytol 218,41-53. |
| [58] | Sakakibara H (2006). Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57,431-449. |
| [59] | Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, Yamaguchi S (2005). Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102,9972-9977. |
| [60] | Sakano Y, Okada Y, Matsunaga A, Suwama T, Kaneko T, Ito K, Noguchi H, Abe I (2004). Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop ( Humulus lupulus L.). Phytochemistry 65,2439-2446. |
| [61] | Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, Yip WK (2007). Nomenclature for two-component signaling elements of rice. Plant Physiol 143,555-557. |
| [62] | Shu K, Liu XD, Xie Q, He ZH (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9,34-45. |
| [63] | Shuai HW, Meng YJ, Luo XF, Chen F, Zhou WG, Dai YJ, Qi Y, Du JB, Yang F, Liu J, Yang WY, Shu K (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci Rep 7,12620. |
| [64] | Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T (2004). Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45,1299-1305. |
| [65] | Steklov MY, Lomin SN, Osolodkin DI, Romanov GA (2013). Structural basis for cytokinin receptor signaling: an evolutionary approach. Plant Cell Rep 32,781-793. |
| [66] | Strnad M (1997). The aromatic cytokinins. Physiol Plant 101,674-688. |
| [67] | Takei K, Sakakibara H, Sugiyama T (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276,26405-26410. |
| [68] | Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007). ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48,263-277. |
| [69] | Tarkowska D, Dole?al K, Tarkowski P, ?stot C, Holub J, Fuksová K, Schmülling T, Sandberg G, Strnad M (2003). Identification of new aromatic cytokinins in Arabidopsis thaliana and Populus × canadensis leaves by LC- (+) ESI-MS and capillary liquid chromatography/frit- fast atom bombardment mass spectrometry. Physiol Plant 117,579-590. |
| [70] | To JPC, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2007). Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19,3901-3914. |
| [71] | Tuan PA, Yamasaki Y, Kanno Y, Seo M, Ayele BT (2019). Transcriptomics of cytokinin and auxin metabolism and signaling genes during seed maturation in dormant and non-dormant wheat genotypes. Sci Rep 9,3983. |
| [72] | Wang YP, Li L, Ye TT, Zhao SJ, Liu Z, Feng YQ, Wu Y (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68,249-261. |
| [73] | Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T (2011). The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol 156,1808-1818. |
| [74] | Wybouw B, De Rybel B (2019). Cytokinin—a developing story. Trends Plant Sci 24,177-185. |
| [75] | Zalaba?k D, Galuszka P, Mrízová K, Podle?a?kova? K, Gu RL, Fre?bortova? J (2014). Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol Biochem 74,283-293. |
| [76] | Zdarska M, Dobisová T, Gelová Z, Pernisová M, Dabravolski S, Hejátko J (2015). Illuminating light, cytokinin, and ethylene signaling crosstalk in plant development. J Exp Bot 66,4913-4931. |
| [77] | Zschiedrich CP, Keidel V, Szurmant H (2016). Molecular mechanisms of two-component signal transduction. J Mol Biol 428,3752-3775. |
| [78] | Zubko E, Adams CJ, Macháèková I, Malbeck J, Scollan C, Meyer P (2002). Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29,797-808. |
| [79] | Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang WJ, Hill K, Raines T, Solano R, Kieber JJ, Loraine AE, Schaller GE (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci USA 114,E5995-E6004. |
| [80] | Zubo YO, Schaller GE (2020). Role of the cytokinin-activated type-B response regulators in hormone crosstalk. Plants 9,166. |
/
| 〈 |
|
〉 |