

Morphology and Genetic Diversity of Phragmites australis in Beijing
Received date: 2020-01-14
Accepted date: 2020-08-26
Online published: 2020-08-26
To understand the distribution and diversity of Phragmites australis in Beijing, we carried out field investigation to Beijing’s major rivers, wetlands and reservoirs, which reveals that the total area of reed populations has reached more than 600 hm 2 in Beijing. The ploidy level is dominated by octaploid, followed by tetraploid. In larger wetlands, the single community of octoploid occupies a dominant position; while in shallow urban rivers, the mixed populations with different morphological, ploidy and genetic diversity are common. There is no significant correlation between the plant traits and ploidy level variation. Six different reed clones were found in Xiaoqing River, all belonging to P haplotypes. The haploid genome size of all clones ranged from (0.499±0.019) pg, with a coefficient of variation of 3.8%. These results show that there is no correlation between phenotype and haplotype. In addition, a reed variant with versicolor leaf characteristics was discovered and named as Phragmites australis var. australis f. Goldstripe. The morphology and genetic diversity of Phragmites in Beijing provide valuable resources for future study of the relationship between reed genotypes and environmental adaptability.
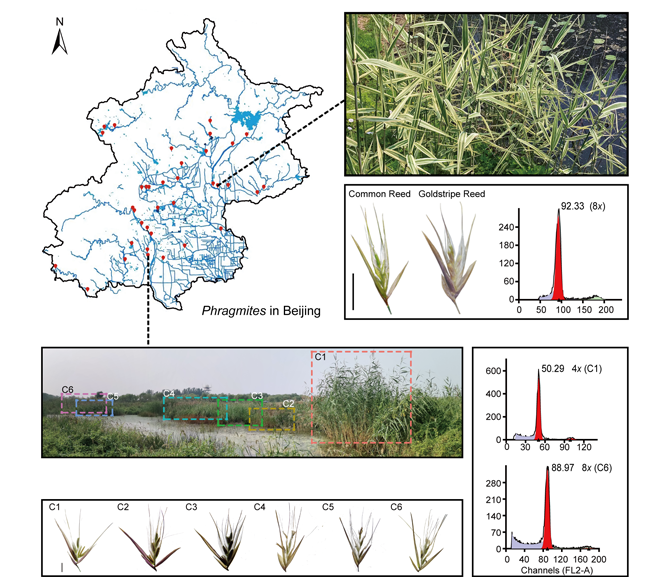
Key words: ploidy level; haplotype; Phragmites australis; polymorphism; variant
Xi Zhang , Tianhang Qiu , Anan Wang , Huajian Zhou , Min Yuan , Li Li , Sulan Bai , Suxia Cui . Morphology and Genetic Diversity of Phragmites australis in Beijing[J]. Chinese Bulletin of Botany, 2020 , 55(6) : 693 -704 . DOI: 10.11983/CBB20006
| [1] | 程杰 (2013). 论中国古代芦苇资源的自然分布、社会利用和文化反映. 阅江学刊 5, 119-134. |
| [2] | 郭春秀, 李发明, 张莹花, 刘淑娟, 朱淑娟, 张大彪 (2012). 河西走廊芦苇草地资源特征及其保护利用. 草原与草坪 32(4), 93-96. |
| [3] | 李建国, 李贵宝, 刘芳, 王殿武, 陈桂珅 (2004). 白洋淀芦苇资源及其生态功能与利用. 南水北调与水利科技 2(5), 37-40. |
| [4] | 张兵 (2008). 谈芦苇湿地的价值. 现代农业科技 (12), 347. |
| [5] | 张承烈, 陈国仓 (1991). 河西走廊不同生态类型芦苇的气体交换特点的研究. 生态学报 11(3), 250-255. |
| [6] | Ahmed MJ (2017). Application of raw and activated Phragmites australis as potential adsorbents for wastewater treatments. Ecol Eng 102, 262-269. |
| [7] | An JX, Wang Q, Yang J, Liu JQ (2012). Phylogeographic analyses of Phragmites australis in China: native distribution and habitat preference of the haplotype that invaded North America. J Syst Evol 50, 334-340. |
| [8] | Burdick DM, Konisky RA (2003). Determinants of expansion for Phragmites australis, common reed, in natural and impacted coastal marshes. Estuaries 26, 407-416. |
| [9] | Clayton WD (1967). Studies in the Gramineae: XIV. Kew Bull 21, 111-117. |
| [10] | Clevering OA, Lissner J (1999). Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquat Bot 64, 185-208. |
| [11] | Cui SX, Hu J, Yang B, Shi L, Huang F, Tsai SN, Ngai SM, He YK, Zhang JH (2009). Proteomic characterization of Phragmites communis in ecotypes of swamp and desert dune. Proteomics 9, 3950-3967. |
| [12] | Cui SX, Wang W, Zhang CL (2002). Plant regeneration from callus cultures in two ecotypes of reed ( Phragmites communis Trinius). In Vitro Cell Dev Biol Plant 38, 325-329. |
| [13] | Dole?el J, Greilhuber J, Suda J (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2, 2233-2244. |
| [14] | Dykyjová D, Pazourková Z (1979). A diploid form of Phragmites communis, as a possible result of cytogenetical response to ecological stress. Folia Geobot Phytotax 14, 113-120. |
| [15] | Eller F, Skálová H, Caplan JS, Bhattarai GP, Burger MK, Cronin JT, Guo WY, Guo X, Hazelton ELG, Kettenring KM, Lambertini C, McCormick MK, Meyerson LA, Mozdzer TJ, Py?ek P, Sorrell BK, Whigham DF, Brix H (2017). Cosmopolitan species as models for ecophysiological responses to global change: the common reed Phragmites australis. Front Plant Sci 8, 1833. |
| [16] | Gorenflot R (1986). Degrés et niveaux de la variation du nombre chromosomique chez Phragmites australis (Cav.) Trin. ex Steud. Ver?ff. Geobot. Inst. ETH, Stiftung Rübel. Zürich 87, 53-65. |
| [17] | Haslam SM (1970). Variation of population type in Phragmites communis Trin. Ann Bot 34, 147-158. |
| [18] | Haslam SM (1971). Community regulation in Phragmites communis Trin. I. monodominant stands. J Ecol 59, 65-73. |
| [19] | Li L, Chen XD, Shi L, Wang CJ, Fu B, Qiu TH, Cui SX (2017). A proteome translocation response to complex desert stress environments in perennial Phragmites sympatric ecotypes with contrasting water availability. Front Plant Sci 8, 511. |
| [20] | Meyerson LA, Cronin JT, Bhattarai GP, Brix H, Lambertini C, Lu?anová M, Rinehart S, Suda J, Py?ek P (2016). Do ploidy level and nuclear genome size and latitude of origin modify the expression of Phragmites australis traits and interactions with herbivores? Biol Invasions 18, 2531-2549. |
| [21] | Meyerson LA, Saltonstall K, Chambers RM, Silliman BR, Bertness MD, Strong D (2009). Phragmites australis in eastern North America: a historical and ecological perspective In: Silliman BR, Grosholz ED, Bertness MD, eds. Human Impacts on Salt Marshes: A Global Perspective. Berkeley: University of California Press. pp. 57-82. |
| [22] | Porebski S, Bailey LG, Baum BR (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15, 8-15. |
| [23] | Raicu P, Staicu S, Stoian V, Roman T (1972). The Phragmites communis Trin. chromosome complement in the Danube delta. Hydrobiologia 39, 83-89. |
| [24] | Saltonstall K (2001). A set of primers for amplification of noncoding regions of chloroplast DNA in the grasses. Mol Ecol Notes 1, 76-78. |
| [25] | Saltonstall K (2002). Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA 99, 2445-2449. |
| [26] | Saltonstall K (2003). Microsatellite variation within and among North American lineages of Phragmites australis. Mol Ecol 12, 1689-1702. |
| [27] | Saltonstall K (2016). The naming of Phragmites haplotypes. Biol Invasions 18, 2433-2441. |
| [28] | Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17, 1105-1109. |
| [29] | Tanaka TST, Irbis C, Inamura T (2017). Phylogenetic analyses of Phragmites spp. in southwest China identified two lineages and their hybrids. Plant Syst Evol 303, 699-707. |
| [30] | Wang WS, Mauleon R, Hu ZQ, Chebotarov D, Tai SS, Wu ZC, Li M, Zheng TQ, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu JL, Sun C, Fu BY, Zhang HL, Gao YM, Zhao XQ, Shen F, Cui X, Yu H, Li ZC, Chen ML, Detras J, Zhou YL, Zhang XY, Zhao Y, Kudrna D, Wang CC, Li R, Jia B, Lu JY, He XC, Dong ZT, Xu JB, Li YH, Wang M, Shi JX, Li J, Zhang DB, Lee S, Hu WS, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu YC, Yue Z, Naredo MEB, Talag J, Wang XQ, Li JJ, Fang XD, Yin Y, Glaszmann JC, Zhang JW, Li JY, Hamilton RS, Wing RA, Ruan J, Zhang GY, Wei CC, Alexandrov N, McNally KL, Li ZK, Leung H (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43-49. |
/
| 〈 |
|
〉 |