

Validation of Rice Candidate Reference Genes for Herbivore-induced Quantitative Real-time PCR Analysis
Received date: 2012-05-22
Revised date: 2012-11-02
Online published: 2013-04-07
Supported by
973
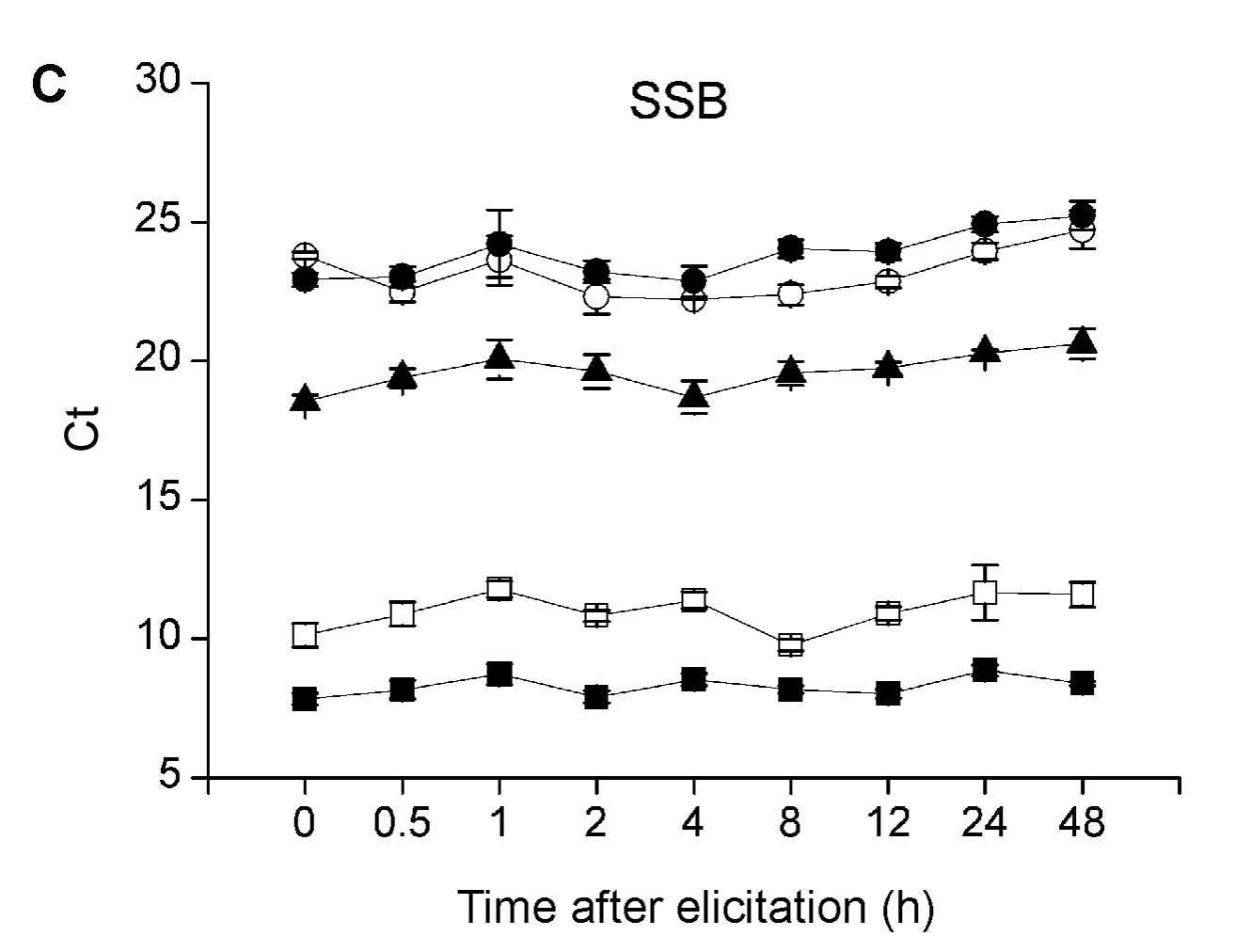
Key words: herbivore-induced; quantitative real-time PCR; reference gene; rice
LI Ran , LI Jian-Cai , ZHOU Guo-Xin , LV Yong-Gen . Validation of Rice Candidate Reference Genes for Herbivore-induced Quantitative Real-time PCR Analysis[J]. Chinese Bulletin of Botany, 2013 , 48(2) : 184 -191 . DOI: 10.3724/SP.J.1259.2013.00184
/
| 〈 |
|
〉 |