

Use of SELEX for Research of the DNA Binding Sites of the RFL, a Transcription Factor of Rice
# Co-first authors
Received date: 2016-06-13
Accepted date: 2016-10-13
Online published: 2017-01-23
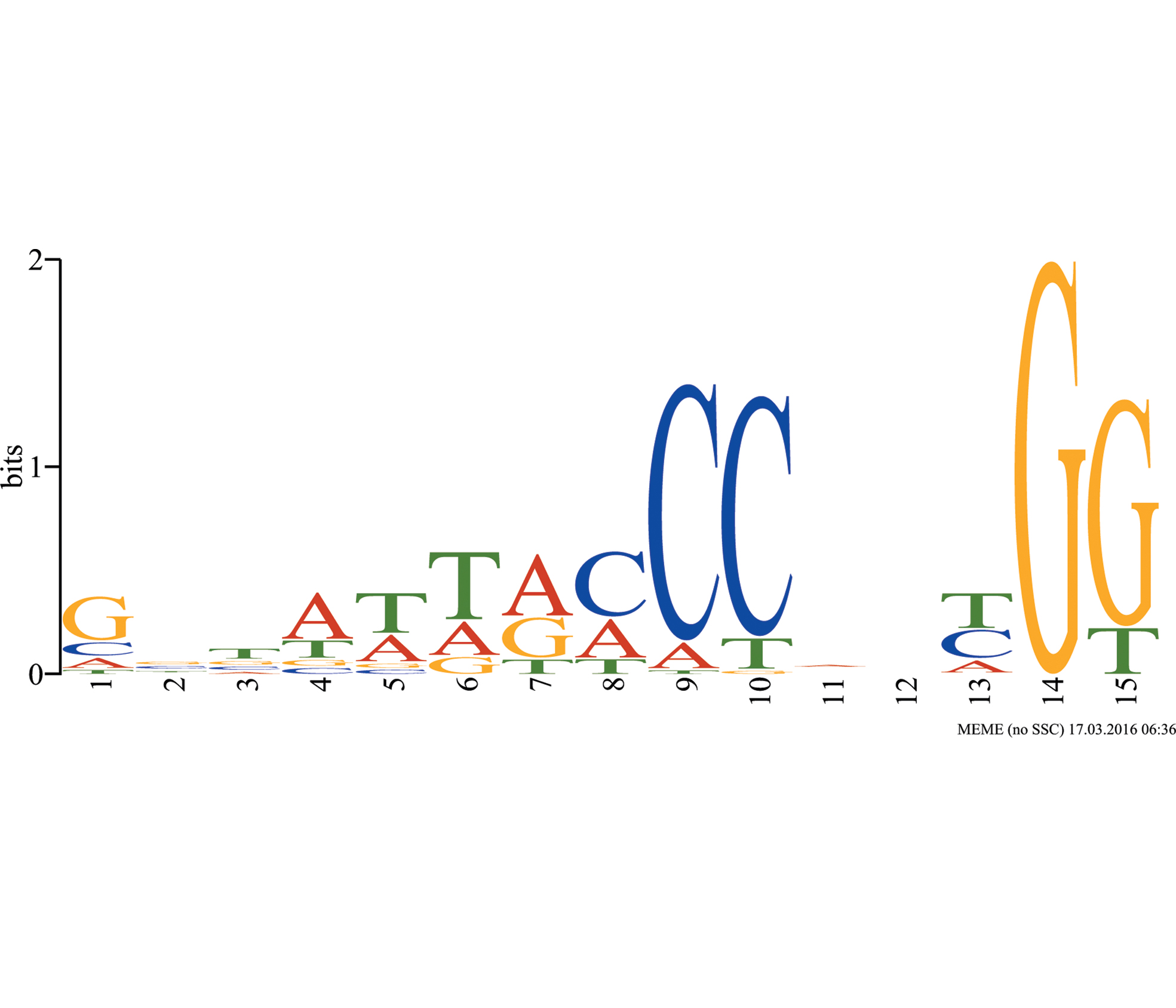
FLORICAULA/LEAFY encodes an evolutionarily conserved land plant-specific transcription factor. It functions by regulating the expression of downstream genes. RFL, the FLORICAULA/LEAFY homolog in rice, controls several important developments in rice: flowering time, plastochron and panicle branching. In this study, we detected the binding specificity of RFL by using the method systematic evolution of ligands by exponential enrichment (SELEX). After seven SELEX processes and sequencing, the binding site of RFL was identified as (C/A)(C/T)NN(T/C/A)G(G/T). The result contributes to clarifying the molecular function of RFL.
Chengqiang Ding , Yanfeng Ding , Shaohua Wang , Zhongyuan Chang , Yan Wang . Use of SELEX for Research of the DNA Binding Sites of the RFL, a Transcription Factor of Rice[J]. Chinese Bulletin of Botany, 2017 , 52(1) : 70 -76 . DOI: 10.11983/CBB16130
| [1] | 马月萍, 陈凡, 戴思兰 (2005). 植物LEAFY同源基因的研究进展. 植物学通报 22, 605-613. |
| [2] | Blazquez MA, Weigel D (2000). Integration of floral inductive signals in Arabidopsis.Nature 404, 889-892. |
| [3] | Busch MA, Bomblies K, Weigel D (1999). Activation of a floral homeotic gene in Arabidopsis.Science 285, 585-587. |
| [4] | Deshpande GM, Ramakrishna K, Chongloi GL, Vijayraghavan U (2015). Functions for rice RFL in vegetative axillary meristem specification and outgrowth.J Exp Bot 66, 2773-2784. |
| [5] | Hames C, Ptchelkine D, Grimm C, Thevenon E, Moyroud E, Gerard F, Martiel JL, Benlloch R, Parcy F, Muller CW (2008). Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins.EMBO J 27, 2628-2637. |
| [6] | Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1.Plant J 69, 168-180. |
| [7] | Klug SJ, Famulok M (1994). All you wanted to know about selex. Mol Biol Rep 20, 97-107. |
| [8] | Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K (1998). Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation.Proc Natl Acad Sci USA 95, 1979-1982. |
| [9] | Pastore JJ, Limpuangthip A, Yamaguchi N, Wu MF, Sang Y, Han SK, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D (2011). LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1.Development 138, 3189-3198. |
| [10] | Prasad K, Kushalappa K, Vijayraghavan U (2003). Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence.Mechan Dev 120, 491-502. |
| [11] | Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008). Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105, 3646-3651. |
| [12] | Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D (2006). The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER.Development 133, 1673-1682. |
| [13] | Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thevenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, Wong GK, Weigel D, Parcy F, Dumas R (2014). A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity.Science 343, 645-648. |
| [14] | Siriwardana NS, Lamb RS (2012). A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana.Plant J 71, 736-749. |
| [15] | Wagner D, Sablowski RW, Meyerowitz EM (1999). Trans- criptional activation of APETALA1 by LEAFY.Science 285, 582-584. |
| [16] | William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004). Genomic identification of direct target genes of LEAFY.Proc Natl Acad Sci USA 101, 1775-1780. |
| [17] | Winter CM, Austin RS, Blanvillain-Baufume S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, Parcy F, Jensen ST, Li H, Wagner D (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response.Dev Cell 20, 430-443. |
/
| 〈 |
|
〉 |