

Received date: 2025-04-01
Accepted date: 2025-06-03
Online published: 2025-06-10
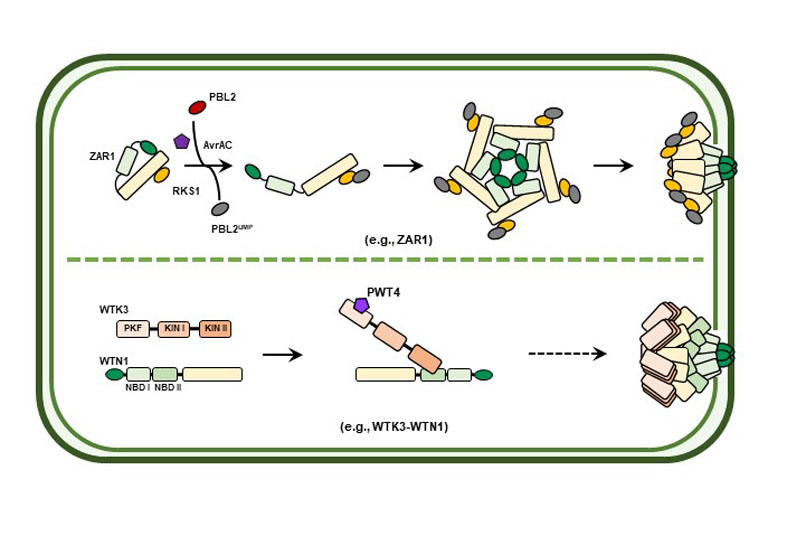
In recent years, we have witnessed transformative breakthroughs in plant disease resistance research, particularly in deciphering the intricate interplay between hosts and pathogens. Cutting-edge discoveries span pathogen recognition mechanisms, immune signaling cascades, and multi-layered interactions integrating plants, pathogens, vectors, and environmental variables. Notably, pioneering studies from domestic research institutions have driven progress across pathogen-sensing systems, secondary metabolite-mediated defense, immune module engineering in crops, and artificial intelligence (AI)-powered solutions for pathogen-resistant peptide design. The rapid development of CRISPR/ Cas9-based gene editing and AI technologies has further empowered researchers to engineer disease-resistant crop varieties with unprecedented precision. Such progress holds profound implications for ensuring national food security and advancing strategic priorities in disease-resistant crop breeding, marking a transformative era in agricultural biotechnology and sustainable agriculture.
Liu Deshui , Yue Ning , Liu Yule . Emerging Innovation in Plant Immunity[J]. Chinese Bulletin of Botany, 2025 , 60(5) : 669 -678 . DOI: 10.11983/CBB25052
| [1] | Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O'Neill M, Reiman D, Tunyasuvunakool K, Wu Z, ?emgulyt? A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, ?ídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493-500. |
| [2] | Ahuja I, Kissen R, Bones AM (2012). Phytoalexins in defense against pathogens. Trends Plant Sci 17, 73-90. |
| [3] | Bednarek P (2012). Chemical warfare or modulators of defence responses—the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15, 407-414. |
| [4] | Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze- Lefert P (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101-106. |
| [5] | Bi GZ, Su M, Li N, Liang Y, Dang S, Xu JC, Hu MJ, Wang JZ, Zou MX, Deng YN, Li QY, Huang SJ, Li JJ, Chai JJ, He KM, Chen YH, Zhou JM (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528-3541. |
| [6] | Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D'Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo NX, Luo MC, Sehgal S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N (2012). Analysis of the bread wheat genome using whole- genome shotgun sequencing. Nature 491, 705-710. |
| [7] | Chen RJ, Chen J, Powell OR, Outram MA, Arndell T, Gajendiran K, Wang YL, Lubega J, Xu Y, Ayliffe MA, Blundell C, Figueroa M, Sperschneider J, Vanhercke T, Kanyuka K, Tang DZ, Zhong GT, Gardener C, Yu GT, Gourdoupis S, Jaremko ?, Matny O, Steffenson BJ, Boshoff WHP, Meyer WB, Arold ST, Dodds PN, Wulff BBH (2025). A wheat tandem kinase activates an NLR to trigger immunity. Science 387, 1402-1408. |
| [8] | Chen SS, Rouse MN, Zhang WJ, Zhang XQ, Guo Y, Briggs J, Dubcovsky J (2020). Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol 225, 948-959. |
| [9] | Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Ara- bidopsis. Science 331, 1185-1188. |
| [10] | Feng F, Zhou JM (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15, 469-476. |
| [11] | F?rderer A, Li ET, Lawson AW, Deng YN, Sun Y, Logemann E, Zhang XX, Wen J, Han ZF, Chang JB, Chen YH, Schulze-Lefert P, Chai JJ (2022). A wheat resistosome defines common principles of immune receptor channels. Nature 610, 532-539. |
| [12] | Graham JH, Bassanezi RB, Dawson WO, Dantzler R (2024). Management of Huanglongbing of citrus: lessons from S?o Paulo and Florida. Ann Rev Phytopathol 62, 243-262. |
| [13] | Halkier BA, Gershenzon J (2006). Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57, 303-333. |
| [14] | Huang SJ, Jia AL, Ma SC, Sun Y, Chang XY, Han ZF, Chai JJ (2023). NLR signaling in plants: from resistosomes to second messengers. Trends Biochem Sci 48, 776-787. |
| [15] | Huang Y, Yang JL, Sun X, Li JH, Cao XQ, Yao SZ, Han YH, Chen CT, Du LL, Li S, Ji YH, Zhou T, Wang H, Han JJ, Wang WM, Wei CH, Xie Q, Yang ZR, Li Y (2025). Perception of viral infections and initiation of antiviral defence in rice. Nature 641, 173-181. |
| [16] | Jones JDG, Dangl JL (2006). The plant immune system. Nature 444, 323-329. |
| [17] | Joubert A, Simoneau P, Campion C, Bataillé-Simoneau N, Iacomi-Vasilescu B, Poupard P, Fran?ois JM, Georgeault S, Sellier E, Guillemette T (2011). Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol Microbiol 79, 1305-1324. |
| [18] | Klymiuk V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen SS, Feng LH, Frenkel Z, Krugman T, Lidzbarsky G, Chang W, J??skel?inen MJ, Schudoma C, Paulin L, Laine P, Bariana H, Sela H, Saleem K, S?rensen CK, Hovm?ller MS, Distelfeld A, Chalhoub B, Dubcovsky J, Korol AB, Schulman AH, Fahima T (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun 9, 3735. |
| [19] | Li SN, Lin DX, Zhang YW, Deng M, Chen YX, Lv B, Li BS, Lei Y, Wang YP, Zhao L, Liang YT, Liu JX, Chen KL, Liu ZY, Xiao J, Qiu JL, Gao CX (2022a). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455-460. |
| [20] | Li ZL, Yang XX, Li WL, Wen ZY, Duan JN, Jiang ZH, Zhang DL, Xie XL, Wang XT, Li FF, Li DW, Zhang YL (2022b). SAMDC3 enhances resistance to Barley stripe mosaic virus by promoting the ubiquitination and proteasomal degradation of viral γb protein. New Phytol 234, 618-633. |
| [21] | Lu P, Guo L, Wang ZZ, Li BB, Li J, Li YH, Qiu D, Shi WQ, Yang LJ, Wang N, Guo GH, Xie JZ, Wu QH, Chen YX, Li MM, Zhang HZ, Dong LL, Zhang PP, Zhu KY, Yu DZ, Zhang Y, Deal KR, Huo NX, Liu CM, Luo MC, Dvorak J, Gu YQ, Li HJ, Liu ZY (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun 11, 680. |
| [22] | Lu P, Zhang GH, Li J, Gong Z, Wang GJ, Dong LL, Zhang HZ, Guo GH, Su M, Wang K, Wang YM, Zhu KY, Wu QH, Chen YX, Li MM, Huang BG, Li BB, Li WL, Dong L, Hou YK, Cui XJ, Fu HK, Qiu D, Yuan CG, Li HJ, Zhou JM, Han GZ, Chen YH, Liu ZY (2025). A wheat tandem kinase and NLR pair confers resistance to multiple fungal pathogens. Science 387, 1418-1424. |
| [23] | Miao P, Wang HJ, Wang W, Wang ZD, Ke H, Cheng HY, Ni JJ, Liang JN, Yao YF, Wang JZ, Zhou JM, Lei XG (2025). A widespread plant defense compound disarms bacterial type III injectisome assembly. Science 387, eads0377. |
| [24] | Mohanty SP, Hughes DP, Salathé M (2016). Using deep learning for image-based plant disease detection. Front Plant Sci 7, 1419. |
| [25] | Ngou BPM, Ahn HK, Ding PT, Jones JDG (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110-115. |
| [26] | Ofer D, Brandes N, Linial M (2021). The language of proteins: NLP, machine learning & protein sequences. Comput Struct Biotechnol J 19, 1750-1758. |
| [27] | Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, García-Casado G, Witters E, Inzé D, Long JA, De Jaeger G, Solano R, Goossens A (2010). NINJA connects the co-repressor TOPLESS to jasmonate signaling. Nature 464, 788-791. |
| [28] | Piasecka A, Jedrzejczak-Rey N, Bednarek P (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206, 948-964. |
| [29] | Reveguk T, Fatiukha A, Potapenko E, Reveguk I, Sela H, Klymiuk V, Li YH, Pozniak C, Wicker T, Coaker G, Fahima T (2025). Tandem kinase proteins across the plant kingdom. Nat Genet 57, 254-262. |
| [30] | Rogers EE, Glazebrook J, Ausubel FM (1996). Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact 9, 748-757. |
| [31] | Sellam A, Dongo A, Guillemette T, Hudhomme P, Simoneau P (2007). Transcriptional responses to exposure to the brassicaceous defence metabolites camalexin and allyl-isothiocyanate in the necrotrophic fungus Alternaria brassicicola. Mol Plant Pathol 8, 195-208. |
| [32] | Sheard LB, Tan X, Mao HB, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, He SY, Rizo J, Howe GA, Zheng N (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co- receptor. Nature 468, 400-405. |
| [33] | Shen QT, Hu T, Bao M, Cao LG, Zhang HW, Song FM, Xie Q, Zhou XP (2016). Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol Plant 9, 911-925. |
| [34] | Shlezinger N, Minz A, Gur Y, Hatam I, Dagdas YF, Talbot NJ, Sharon A (2011). Anti-apoptotic machinery protects the necrotrophic fungus Botrytis cinerea from host-induced apoptotic-like cell death during plant infection. PLoS Pathog 7, e1002185. |
| [35] | Tang XY, Xiao YM, Zhou JM (2006). Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact 19, 1159-1166. |
| [36] | Tian HN, Wu ZS, Chen SY, Ao K, Huang WJ, Yaghmaiean H, Sun TJ, Xu F, Zhang YJ, Wang SC, Li X, Zhang YL (2021). Activation of TIR signaling boosts pattern-triggered immunity. Nature 598, 500-503. |
| [37] | Vargas P, Farias GA, Nogales J, Prada H, Carvajal V, Barón M, Rivilla R, Martín M, Olmedilla A, Gallegos MT (2013). Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ Microbiol Rep 5, 841-850. |
| [38] | Vargas P, Felipe A, Michán C, Gallegos MT (2011). Induction of Pseudomonas syringae pv. tomato DC3000 MexAB-OprM multidrug efflux pump by flavonoids is mediated by the repressor PmeR. Mol Plant Microbe Interact 24, 1207-1219. |
| [39] | Wang JZ, Hu MJ, Wang J, Qi JF, Han ZF, Wang GX, Qi YJ, Wang HW, Zhou JM, Chai JJ (2019a). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870. |
| [40] | Wang JZ, Wang J, Hu MJ, Wu S, Qi JF, Wang GX, Han ZF, Qi YJ, Gao N, Wang HW, Zhou JM, Chai JJ (2019b). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868. |
| [41] | Wang W, Yang J, Zhang J, Liu YX, Tian CP, Qu BY, Gao CL, Xin PY, Cheng SJ, Zhang WJ, Miao P, Li L, Zhang XJ, Chu JF, Zuo JR, Li JY, Bai Y, Lei XG, Zhou JM (2020). An Arabidopsis secondary metabolite directly targets expression of the bacterial type III secretion system to inhibit bacterial virulence. Cell Host Microbe 27, 601-613. |
| [42] | Wang Y, Pruitt RN, Nürnberger T, Wang YC (2022). Evasion of plant immunity by microbial pathogens. Nat Rev Microbiol 20, 449-464. |
| [43] | Wang YJ, Gong Q, Wu YY, Huang F, Ismayil A, Zhang DF, Li HG, Gu HQ, Ludman M, Fátyol K, Qi YJ, Yoshioka K, Hanley-Bowdoin L, Hong YG, Liu YL (2021). A calmodulin-binding transcription factor links calcium signaling to antiviral RNAi defense in plants. Cell Host Microbe 29, 1393-1406. |
| [44] | Washburn JD, Mejia-Guerra MK, Ramstein G, Kremling KA, Valluru R, Buckler ES, Wang H (2019). Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence. Proc Natl Acad Sci USA 116, 5542-5549. |
| [45] | Wu DW, Qi TC, Li WX, Tian HX, Gao H, Wang JJ, Ge J, Yao RF, Ren CM, Wang XB, Liu YL, Kang L, Ding SW, Xie DX (2017a). Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res 27, 402-415. |
| [46] | Wu JG, Yang RX, Yang ZR, Yao SZ, Zhao SS, Wang Y, Li PC, Song XW, Jin L, Zhou T, Lan Y, Xie LH, Zhou XP, Chu CC, Qi YJ, Cao XF, Li Y (2017b). ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants 3, 16203. |
| [47] | Wu JG, Yang ZR, Wang Y, Zheng LJ, Ye RQ, Ji YH, Zhao SS, Ji SY, Liu RF, Xu L, Zheng H, Zhou YJ, Zhang X, Cao XF, Xie LH, Wu ZJ, Qi YJ, Li Y (2015). Viral-inducible argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4, e05733. |
| [48] | Wu JG, Zhang YL, Li FF, Zhang XM, Ye J, Wei TY, Li ZH, Tao XR, Cui F, Wang XB, Zhang LL, Yan F, Li SF, Liu YL, Li DW, Zhou XP, Li Y (2024). Plant virology in the 21st century in China: recent advances and future directions. J Integr Plant Biol 66, 579-622. |
| [49] | Xin XF, Kvitko B, He SY (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16, 316-328. |
| [50] | Yang ZR, Huang Y, Yang JL, Yao SZ, Zhao K, Wang DH, Qin QQ, Bian Z, Li Y, Lan Y, Zhou T, Wang H, Liu C, Wang WM, Qi YJ, Xu ZH, Li Y (2020). Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89-103. |
| [51] | Yuan MH, Jiang ZY, Bi GZ, Nomura K, Liu MH, Wang YP, Cai BY, Zhou JM, He SY, Xin XF (2021). Pattern- recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105-109. |
| [52] | Zhao PZ, Yang H, Sun YW, Zhang JY, Gao KX, Wu JB, Zhu CR, Yin CC, Chen XY, Liu Q, Xia QD, Li Q, Xiao H, Sun HX, Zhang XX, Yi L, Zhou CY, Kliebenstein DJ, Fang RX, Wang XF, Ye J (2025). Targeted MYC2 stabilization confers citrus Huanglongbing resistance. Science 388, 191-198. |
| [53] | Zhou JM, Zhang YL (2020). Plant immunity: danger perception and signaling. Cell 181, 978-989. |
| [54] | Zhou WK, Lozano-Torres JL, Blilou I, Zhang XY, Zhai QZ, Smant G, Li CY, Scheres B (2019). A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942-956. |
/
| 〈 |
|
〉 |