

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (4): 522-530.DOI: 10.11983/CBB18229 cstr: 32102.14.CBB18229
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Received:2018-10-30
Accepted:2019-02-11
Online:2019-07-01
Published:2020-01-08
Contact:
Jiwei Ruan
Fan Li,Jiwei Ruan. High-throughput Identification of Meiotic Anti-CO Mutants by Fluorescent Reporters[J]. Chinese Bulletin of Botany, 2019, 54(4): 522-530.
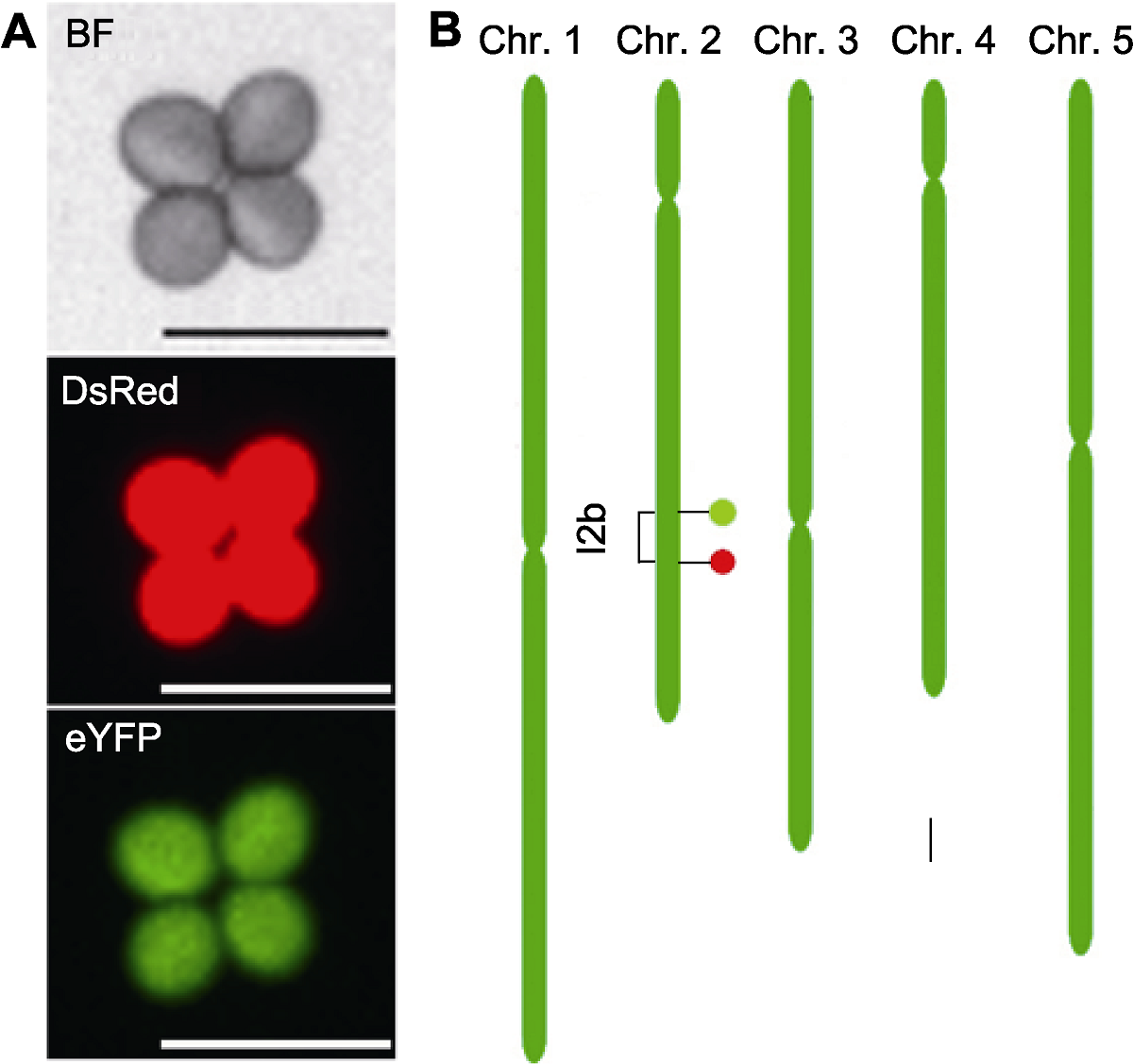
Figure 1 A schematic illustration of Arabidopsis fluorescent tagged line FTL-I2b (A) Fluorescence micrograph of a tetrad pollen of FTL-I2b, which contains a DsRed and an eYFP fluorescent marker and expresses red fluorescence and green fluorescence under different fluorescence excitations respectively (Bars=50 μm); (B) The genomic location of FTL-I2b fluorescent markers on the chromosome (Bar=1 Mb), the DsRed and eYFP fluorescent markers are indicated by filled circles colored by red (down) and green (up), respectively, which constructing a I2b interval (1.45 Mb). BF: Bright field
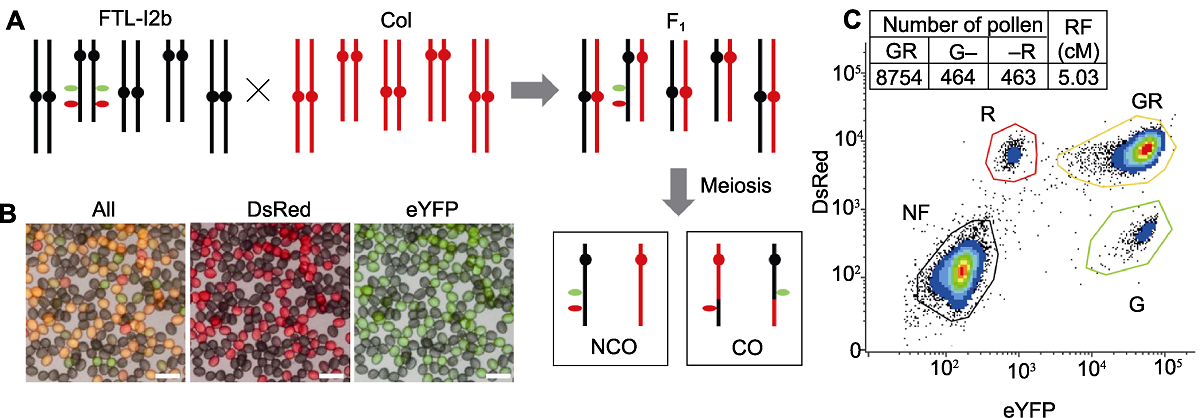
Figure 2 The Arabidopsis hemizygous fluorescent markers (I2b/++) of FTL-I2b (A) A schematic illustration of FTL-I2b cross with Col, the hybrid F1 contains a hemizygous fluorescent markers after hybridization, the recombined and unrecombined gametes are formed through meiosis; (B) Fluorescence micrograph of pollen in hybrid F1 generation, in which yellow pollen (GR) is unrecombined gamete, and monochromatic red (R) and green (G) pollen is recombined gametes (Bars=50 μm); (C) The recombination frequency (RF) of I2b interval can be calculated by counting the fluorescent marker pattern in the gametes via flow cytometry, the formula calculating recombination frequency=(G+R)/(2GR+G+R)×100.
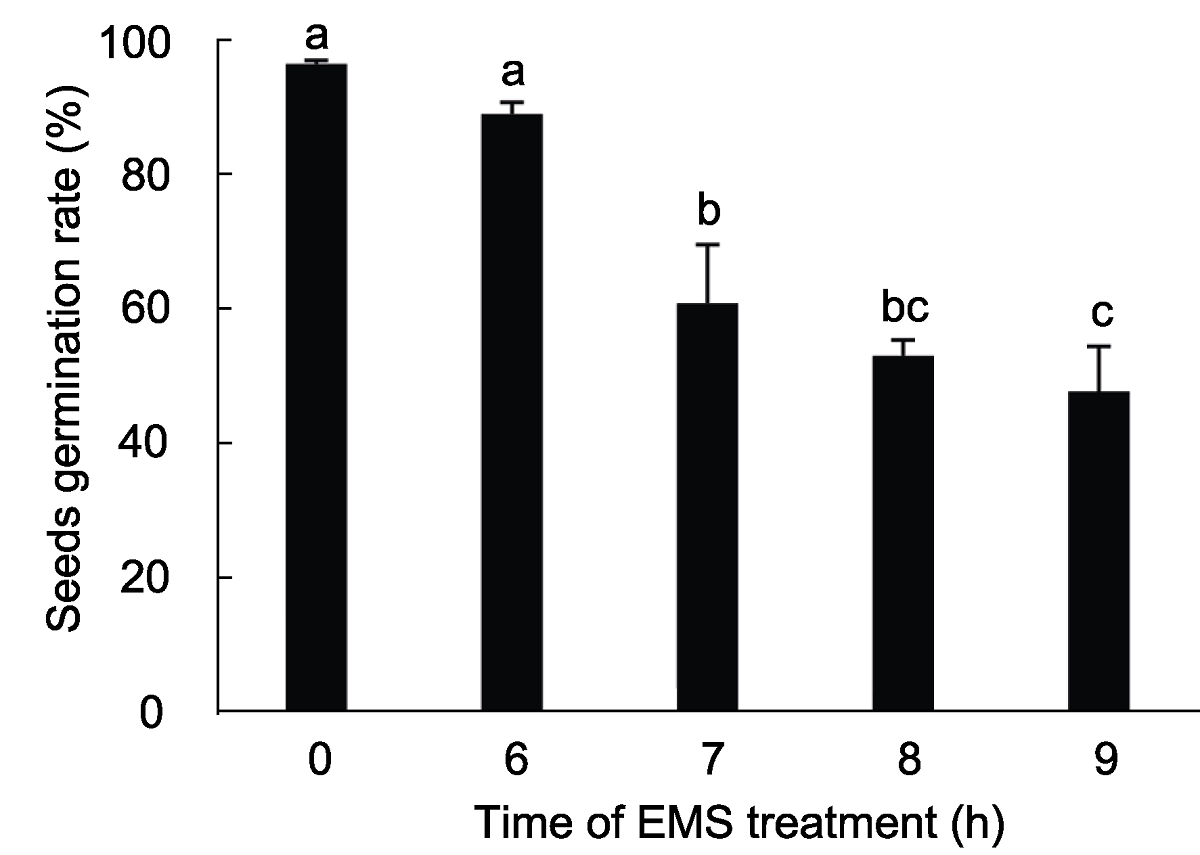
Figure 3 The germination rate of M1 Arabidopsis seeds by different treat time using 75 mmol∙L-1 EMS The average germination rate was derived from three repeats, different lowercase letters indicate significant differences (P<0.05).
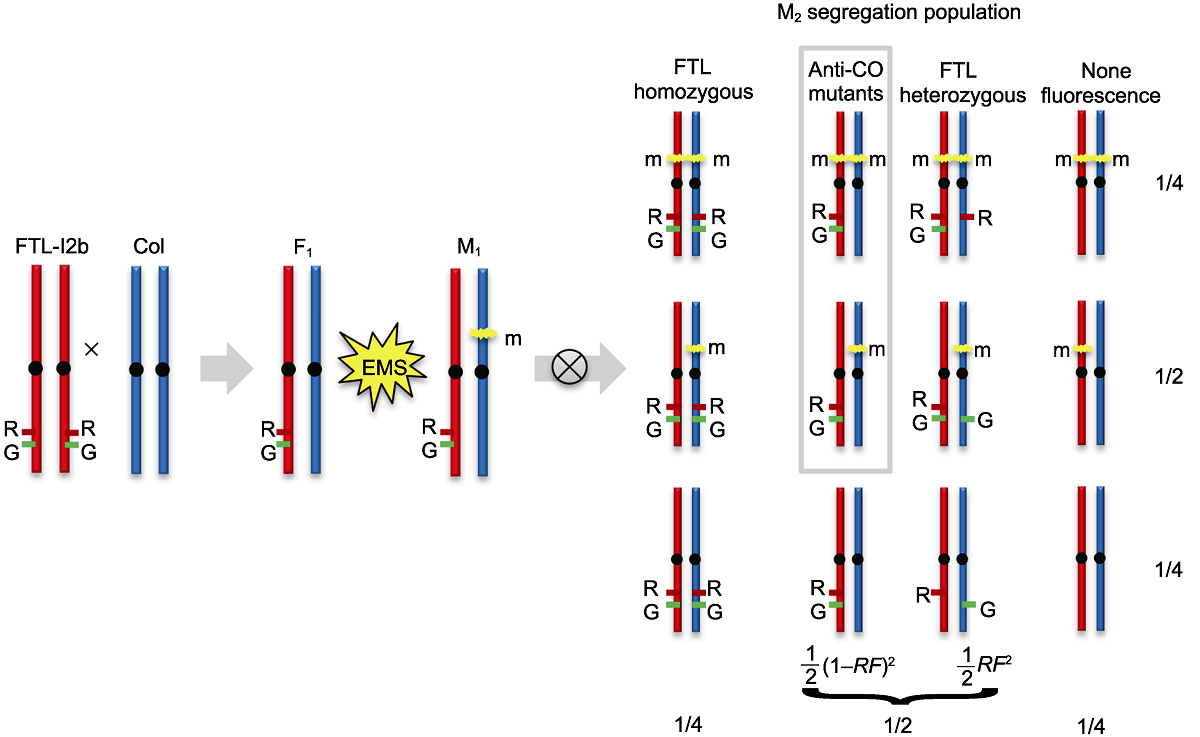
Figure 4 A schematic illustration of Arabidopsis meiotic anti-CO mutants forward genetics screen The forward genetics mutants screen was used the FTL-I2b containing linked hemizygous fluorescent reporters (I2b/++) on the genetic background of Arabidopsis wild type (WT). High-throughput detecting the recombination frequency by flow cytometry could identify the mutants with increased recombination rate. The DsRed and eYFP fluorescent reporters of FTL-I2b are indicated by R and G, respectively. The m refers to a dominant or recessive mutation. RF: Recombination frequency
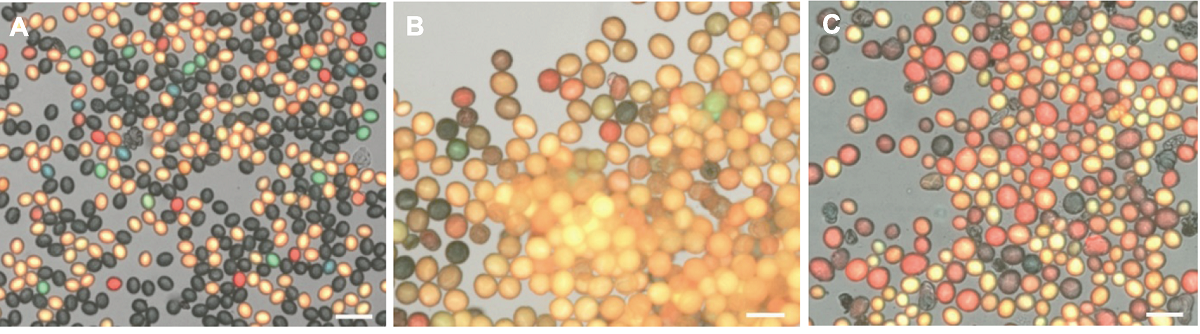
Figure 5 The pollen micrograph of Arabidopsis wild type and big pollen mutants (A) The pollen size of Arabidopsis wild type (WT) is (21.82±0.98) µm (n=10); (B) The large pollen rate of big pollen 1 mutant is 100%, pollen size is (38.36±1.37) µm (n=10); (C) The large pollen rate of big pollen 2 mutant is 50%, pollen size is (36.00±2.70) µm (n=10). Bars=50 µm

Figure 6 Arabidopsis wild type and recombination suppressor mutants (A) The plant phenotype of Arabidopsis wild type (WT), dominant recombination suppressor mutant (drs) and recessive recombination suppressor mutant (rrs1) (Bars=1 cm); (B) The fluorescent pollen micrograph of Arabidopsis wild type (WT), dominant recombination suppressor mutant (drs) and recessive recombination suppressor mutant (rrs1) (Bars=50 μm)
| [1] | Berchowitz LE, Copenhaver GP ( 2008). Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3, 41-50. |
| [2] | Berchowitz LE, Copenhaver GP ( 2010). Genetic interference: don't stand so close to me. Curr Genomics 11, 91-102. |
| [3] | Berchowitz LE, Francis KE, Bey AL, Copenhaver GP ( 2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3, e132. |
| [4] | Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R ( 2012). FANCM limits meiotic crossovers. Science 336, 1588-1590. |
| [5] | Fernandes JB, Duhamel M, Seguéla-Arnaud M, Froger N, Girard C, Choinard S, Solier V, De Winne N, De Jaeger G, Gevaert K, Andrey P, Grelon M, Guerois R, Kumar R, Mercier R ( 2017). FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet 14, e1007317. |
| [6] | Fernandes JB, Séguéla-Arnaud M, Larchevêque C, Lloyd AH, Mercier R ( 2018). Unleashing meiotic crossovers in hybrid plants. Proc Natl Acad Sci USA 115, 2431-2436. |
| [7] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP ( 2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [8] | Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lehmemdi A, Mazel J, Crismani W, Mercier R ( 2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet 11, e1005369. |
| [9] | Girard C, Crismani W, Froger N, Mazel J, Lemhemdi A, Horlow C, Mercier R ( 2014). FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res 42, 9087-9095. |
| [10] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C ( 2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [11] | Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, Sekelsky J ( 2017). Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol 27, 96-102. |
| [12] | Higgins JD, Buckling EF, Franklin FC, Jones GH ( 2008). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54, 152-162. |
| [13] | Hu Q, Li YF, Wang HJ, Shen Y, Zhang C, Du GJ, Tang D, Cheng ZK ( 2017). Meiotic chromosome association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29, 1697-1708. |
| [14] | Huang JY, Cheng ZH, Wang C, Hong Y, Su H, Wang J, Copenhaver GP, Ma H, Wang YX ( 2015). Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc Natl Acad Sci USA 112, 12534-12539. |
| [15] | Jones GH, Franklin FCH ( 2006). Meiotic crossing-over: obligation and interference. Cell 126, 246-248. |
| [16] | Kim Y, Schumaker KS, Zhu JK ( 2006). EMS mutagenesis of Arabidopsis. In: Salinas J, Sanchez-Serrano JJ, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 101-103. |
| [17] | Kurzbauer MT, Pradillo M, Kerzendorfer C, Sims J, Ladurner R, Oliver C, Janisiw MP, Mosiolek M, Schweizer D, Copenhaver GP, Schlögelhofer P ( 2018). Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30, 415-428. |
| [18] | Li F, De Storme N, Geelen D ( 2017). Dynamics of male meiotic recombination frequency during plant development using fluorescent tagged lines in Arabidopsis thaliana. Sci Rep 7, 42535. |
| [19] | Lu PL, Han XW, Qi J, Yang JG, Wijeratne AJ, Li T, Ma H ( 2012). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res 22, 508-518. |
| [20] | Lu PL, Wijeratne AJ, Wang ZJ, Copenhaver GP, Ma H ( 2014). Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions. J Genet Genomics 41, 165-175. |
| [21] | Lukowitz W, Gillmor CS, Scheible WR ( 2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123, 795-806. |
| [22] | Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R ( 2008). SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18, 1432-1437. |
| [23] | Macaisne N, Vignard J, Mercier R ( 2011). SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124, 2687-2691. |
| [24] | Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M ( 2015). The molecular biology of meiosis in plants. Annu Rev Plant Biol 66, 297-327. |
| [25] | Mieulet D, Aubert G, Bres C, Klein A, Droc G, Vieille E, Rond-Coissieux C, Sanchez M, Dalmais M, Mauxion JP, Rothan C, Guiderdoni E, Mercier R ( 2018). Unleashing meiotic crossovers in crops. Nat Plants 4, 1010-1016. |
| [26] | Qi J, Chen YM, Copenhaver GP, Ma H ( 2014). Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc Natl Acad Sci USA 111, 10007-10012. |
| [27] | Qu LJ, Qin GJ ( 2014). Generation and identification of Arabidopsis EMS mutants. In: Sanchez-Serrano JJ, Salinas J, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 225-239. |
| [28] | Séguéla-Arnaud M, Choinard S, Larchevêque C, Girard C, Froger N, Crismani W, Mercier R ( 2017). RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res 4, 1860-1871. |
| [29] | Séguéla-Arnaud M, Crismani W, Larchevêque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K, De Jaeger G, Chelysheva L, Mercier R ( 2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA 112, 4713-4718. |
| [30] | Wang YX, Cheng ZH, Huang JY, Shi Q, Hong Y, Copenhaver GP, Gong ZZ, Ma H ( 2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet 8, e1003039. |
| [31] | Yelina NE, Lambing C, Hardcastle TJ, Zhao XH, Santos B, Henderson IR ( 2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Gene Dev 29, 2183-2202. |
| [32] | Yelina NE, Ziolkowski PA, Miller N, Zhao X, Kelly KA, Muñoz DF, Mann DJ, Copenhaver GP, Henderson IR ( 2013). High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat Protoc 8, 2119-2134. |
| [1] | Xinjie Cheng, Hengxiu Yu, Zhukuan Cheng. Protocols for Analyzing Rice Meiotic Chromosomes [J]. Chinese Bulletin of Botany, 2019, 54(4): 503-508. |
| [2] | Zhihui Xue, Kang Chong. Chinese Scientists Make Groundbreaking Discoveries in Clonal Propagation of F1 Hybrids [J]. Chinese Bulletin of Botany, 2019, 54(1): 1-3. |
| [3] | Xiangdong Luo, Liangfang Dai, Yong Wan, Biaolin Hu, Fosheng Li, Xia Li, Jiankun Xie. Cytological Studies of Male Gametogenesis and Development in the Reciprocal Interspecific Hybrid F1 of Dongxiang Wild Rice (Oryza rufipogon) and Oryza sativa [J]. Chinese Bulletin of Botany, 2011, 46(4): 407-412. |
| [4] | Hexin Tan;Tieqiao Wen;Dabing Zhang. Molecular Mechanisms of Pollen Development in Oryza sativa [J]. Chinese Bulletin of Botany, 2007, 24(03): 330-339. |
| [5] | LU Bao-Rong, Bjφrn Salomon. Differentiation of the StY genomes in Elymus species as referred by meiotic pairing in interspecific hybrids and its evolutionary significance [J]. Biodiv Sci, 2004, 12(2): 213-226. |
| [6] | GUO Jun-Yang CHEN Jin-Feng② QIAN Chun-Tao CAO Qing-He. Meiotic Chromosome Pairing Research and Genome Analysis in Plants [J]. Chinese Bulletin of Botany, 2004, 21(05): 513-520. |
| [7] | LI Ya-Xuan. Advances in the Research on Plant Meiosis [J]. Chinese Bulletin of Botany, 1999, 16(05): 526-529. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
