

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (4): 490-499.DOI: 10.11983/CBB22080 cstr: 32102.14.CBB22080
• INVITED PROTOCOL • Previous Articles Next Articles
Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei
Received:2022-04-12
Revised:2022-06-23
Online:2022-07-01
Published:2022-07-14
About author:First author contact:† These authors contributed equally to this paper
Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei. An Effective in Vitro SUMOylation Detection System for Plant Proteins[J]. Chinese Bulletin of Botany, 2022, 57(4): 490-499.
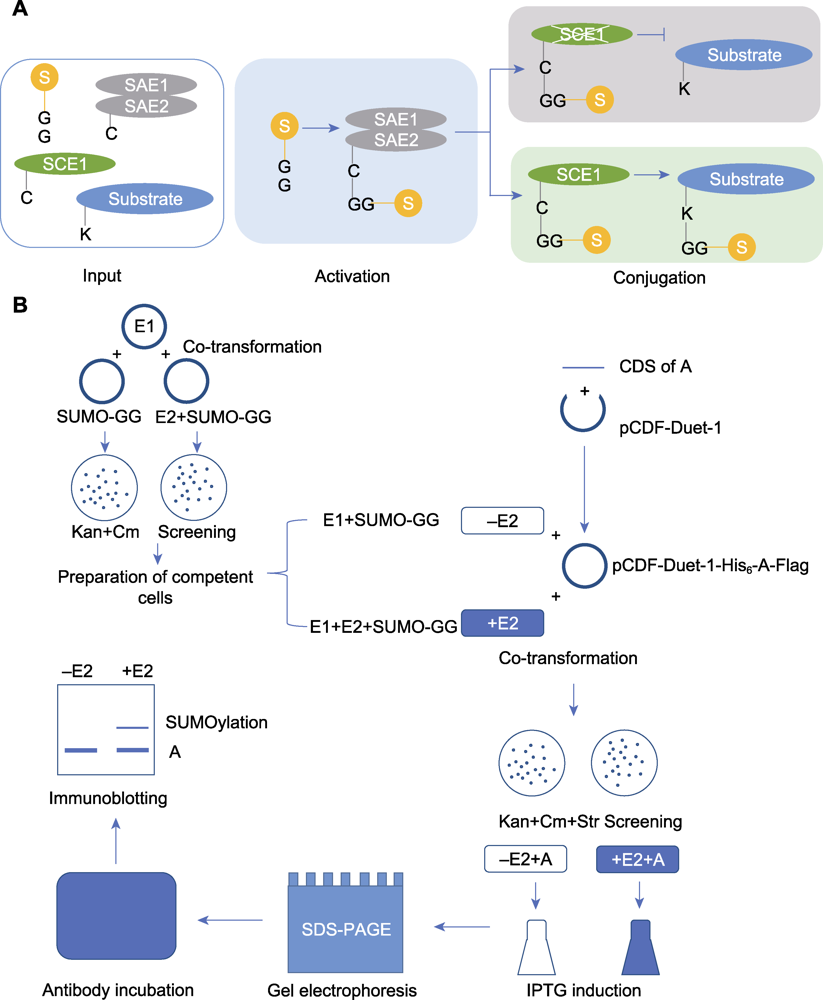
Figure 1 The process of in vitro SUMOylation analysis (A) The process diagram of in vitro SUMOylation detection (Input: the key component genes of SUMO modification and the candidate substrate protein gene were introduced into Escherichia coli for expression; Activating: E1 activates SUMO molecules to participate in the modification; Conjugating: E2 attaches the activated SUMO molecules to the substrate); (B) The experimental procedure of in vitro SUMOylation detection (Co-transformation: The plasmids for expressing key SUMOylation components were introduced into E. coli, and the positive colonies were screened on LB agar plates containing kanamycin and chloramphenicol; Preparation of competent cells: The obtained positive colonies were prepared into competent cells; Co-transformation: The constructed plasmid for substrate protein expression was introduced into the competent cells mentioned above, and positive colonies were screened on LB agar plates by kanamycin, chloramphenicol and streptomycin; Induction: Protein expression was induced by IPTG; Gel electrophoresis: After lysed and denatured, samples were separated by SDS-PAGE. Then the proteins were transferred from gel to PVDF membrane; Antibody incubation: The blocked PVDF membrane was incubated with antibody; Immunoblotting: Observation and recording of the experimental results)
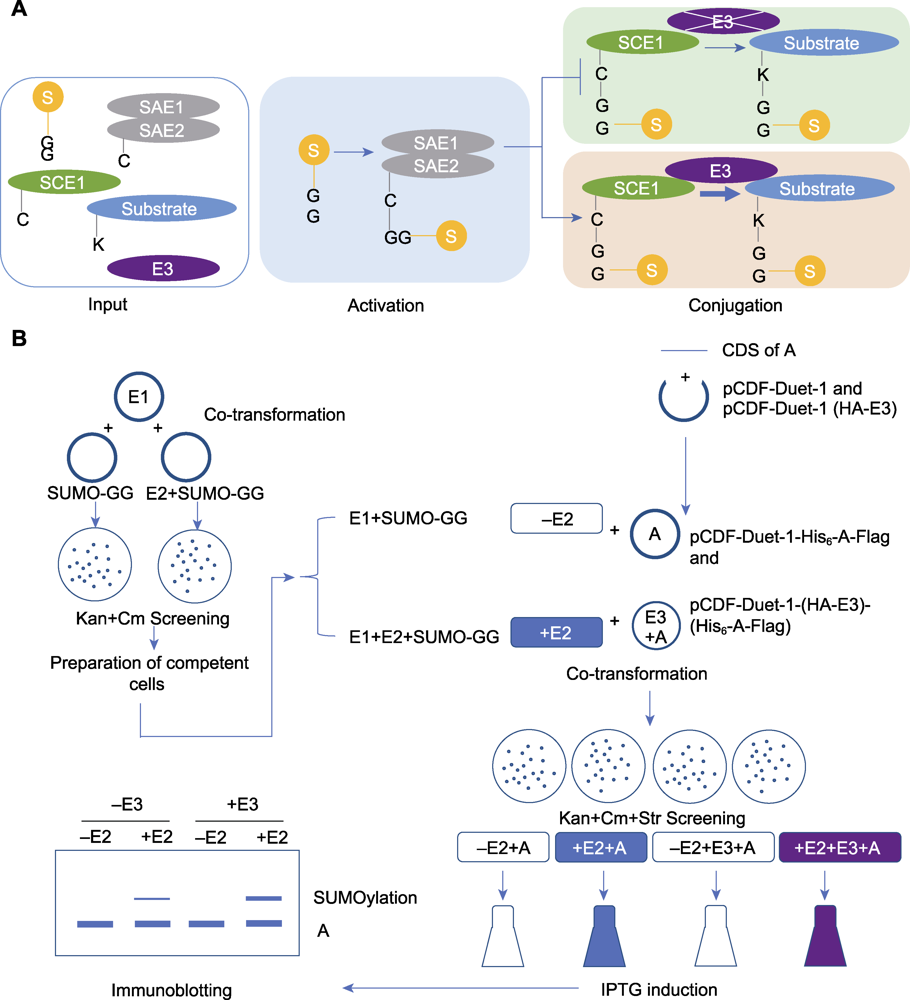
Figure 2 The process of in vitro SUMOylation analysis mediated by an E3 ligase (A) The process diagram of the E3 ligase-mediated in vitro SUMOylation detection (Input: The key component genes of SUMO modification and the candidate substrate protein gene were introduced into Escherichia coli for expression; Activating: E1 activates SUMO molecules to participate in the modification; Conjugating: E2 attaches the activated SUMO molecules to the substrate. In the presence of E3, E3 facilitates the attachment of SUMO molecules from E2 to the substrate protein); (B) The experimental procedure of the E3 ligase-mediated in vitro SUMOylation detection (Co-transformation: The plasmids for expression of key SUMOylation components were introduced into E. coli, and the positive colonies were screened on LB agar plates containing kanamycin and chloramphenicol; Preparation of competent cells: The obtained positive colonies were transformed into competent cells; Co-transformation: The constructed plasmid for substrate protein expression (with independent E3 expression cassette) was introduced into the competent cells mentioned above, and the positive colonies were screened by LB agar plates containing kanamycin, chloramphenicol and streptomycin; Induction: Protein expression was induced by IPTG; Immunoblotting: After lysed and denatured, samples were separated by SDS-PAGE gel electrophoresis, then the proteins were transferred from gel to PVDF membrane, the blocked PVDF membrane was incubated with antibody, the result was observed and recorded by immunoblotting).
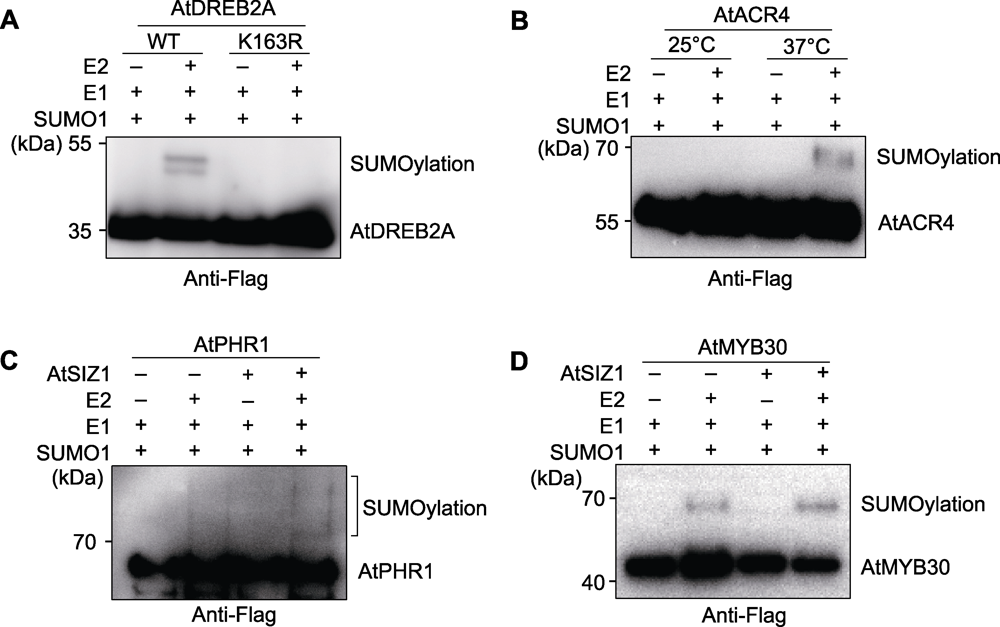
Figure 3 Experimental cases (A) SUMOylation test of AtDREB2A; (B) Temperature affects SUMOylation of AtACR4; (C) The E3 ligase AtSIZ1 facilitates SUMOylation of AtPHR1; (D) The E3 ligase AtSIZ1 facilitates SUMOylation of AtMYB30
| [1] |
韩丹璐, 赖建彬, 阳成伟 (2018). SUMO E3连接酶在植物生长发育中的功能研究进展. 植物学报 53, 175-184.
DOI |
| [2] |
曲高平, 金京波 (2020). 植物蛋白SUMO化修饰检测方法. 植物学报 55, 83-89.
DOI |
| [3] |
Augustine RC, Vierstra RD (2018). SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol 45, 143-154.
DOI PMID |
| [4] | Dai Vu L, Gevaert K, De Smet I (2018). Protein language: post-translational modifications talking to each other. Tren- ds Plant Sci 23, 1068-1080. |
| [5] |
Fang Q, Zhang J, Zhang Y, Fan N, van den Burg HA, Huang CF (2020). Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32, 3921-3938.
DOI URL |
| [6] |
Han DL, Lai JB, Yang CW (2021). SUMOylation: a critical transcription modulator in plant cells. Plant Sci 310, 110987.
DOI URL |
| [7] |
Huang LX, Yang SG, Zhang SC, Liu M, Lai JB, Qi YL, Shi SF, Wang JX, Wang YQ, Xie Q, Yang CW (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60, 666-678.
DOI URL |
| [8] |
Jiang JM, Xie Y, Du JJ, Yang CW, Lai JB (2021). A SUMO ligase OsMMS21 regulates rice development and auxin response. J Plant Physiol 263, 153447.
DOI URL |
| [9] |
Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53, 530-540.
DOI URL |
| [10] |
Kong XF, Hong YC, Hsu YF, Huang H, Liu X, Song Z, Zhu JK (2020). SIZ1-mediated SUMOylation of ROS1 enhances its stability and positively regulates active DNA demethylation in Arabidopsis. Mol Plant 13, 1816-1824.
DOI URL |
| [11] |
Li YR, Williams B, Dickman M (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)- mediated heat tolerance requires translocation, SUMOylation and binding to WRKY29. New Phytol 214, 695-705.
DOI URL |
| [12] |
Liu YY, Lai JB, Yu MY, Wang FG, Zhang JJ, Jiang JM, Hu H, Wu Q, Lu GH, Xu PL, Yang CW (2016). The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/ DPa complex in cell cycle regulation. Plant Cell 28, 2225-2237.
DOI URL |
| [13] |
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007). SIZ1-mediated SUMOylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403-1414.
DOI URL |
| [14] |
Miura K, Lee J, Miura T, Hasegawa PM (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51, 103-113.
DOI URL |
| [15] |
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102, 7760-7765.
DOI URL |
| [16] |
Morrell R, Sadanandom A (2019). Dealing with stress: a review of plant SUMO proteases. Front Plant Sci 10, 1122.
DOI URL |
| [17] |
Niu D, Lin XL, Kong XX, Qu GP, Cai B, Lee J, Jin JB (2019). SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12, 215-228.
DOI PMID |
| [18] |
Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama JI, Tanaka K (2009). Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50, 1049-1061.
DOI URL |
| [19] |
Perry JJP, Tainer JA, Boddy MN (2008). A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33, 201-208.
DOI URL |
| [20] |
Roy D, Sadanandom A (2021). SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants. Cell Mol Life Sci 78, 2641-2664.
DOI URL |
| [21] |
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress- responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103, 18822-18827.
DOI URL |
| [22] |
Saleh A, Withers J, Mohan R, Marqués J, Gu YN, Yan SP, Zavaliev R, Nomoto M, Tada Y, Dong XN (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169-182.
DOI URL |
| [23] |
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145, 119-134.
PMID |
| [24] | Verma V, Srivastava AK, Gough C, Campanaro A, Srivastava M, Morrell R, Joyce J, Bailey M, Zhang CJ, Krysan PJ, Sadanandom A (2021). SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc Natl Acad Sci USA 118, e2021351118. |
| [25] | Wang FG, Liu YY, Shi YQ, Han DL, Wu YY, Ye WX, Yang HL, Li GW, Cui F, Wan SB, Lai JB, Yang CW (2020). SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant Physiol 183, 41-50. |
| [26] |
Wang Z, Prelich G (2009). Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 29, 1694-1706.
DOI URL |
| [27] |
Xu JM, Zhu JY, Liu JJ, Wang JX, Ding ZJ, Tian HY (2021). SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis. J Integr Plant Biol 63, 1147-1160.
DOI URL |
| [28] |
Zhang JJ, Lai JB, Wang FG, Yang SG, He ZP, Jiang JM, Li QL, Wu Q, Liu YY, Yu MY, Du JJ, Xie Q, Wu KQ, Yang CW (2017). A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant Physiol 173, 1574-1582.
DOI URL |
| [29] |
Zheng Y, Schumaker KS, Guo Y (2012). SUMOylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109, 12822-12827.
DOI URL |
| [30] |
Zhou LJ, Zhang CL, Zhang RF, Wang GL, Li YY, Hao YJ (2019). The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis. Plant Physiol 179, 88-106.
DOI PMID |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||