

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (1): 98-110.DOI: 10.11983/CBB21175 cstr: 32102.14.CBB21175
• SPECIAL TOPICS • Previous Articles Next Articles
Lingyu Ma1,3†, Xiaohong Qi1†, Zijian Hu1,2, Weiwei Shen1,2, Guangchao Wang1,2, Bolin Zhang1,2,4, Xi Zhang1,2,4,*( ), Jinxing Lin1,2,4,*(
), Jinxing Lin1,2,4,*( )
)
Received:2021-10-11
Accepted:2022-01-13
Online:2022-01-01
Published:2022-01-17
Contact:
Xi Zhang,Jinxing Lin
About author:First author contact:†These authors contributed equally to this paper
Lingyu Ma, Xiaohong Qi, Zijian Hu, Weiwei Shen, Guangchao Wang, Bolin Zhang, Xi Zhang, Jinxing Lin. Applications of Optical Clearing Technique in Multi-scale Imaging[J]. Chinese Bulletin of Botany, 2022, 57(1): 98-110.
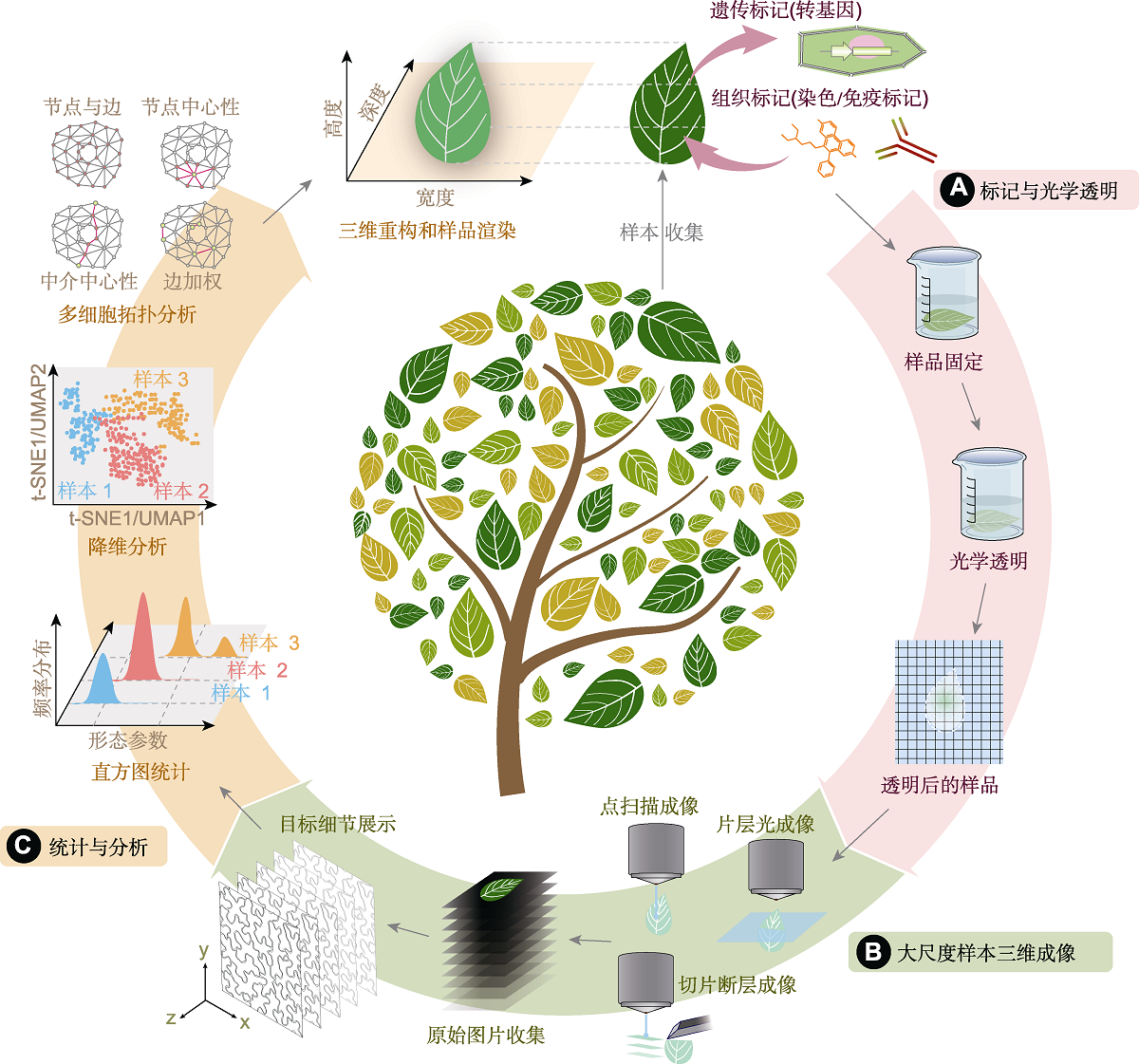
Figure 1 Workflow of optical clearing, three-dimensional (3D) imaging and statistical analysis of plant sample (A) Labeling and optical clearing of plant sample, mainly including genetic labeling (transgene) and histological labeling (immunolabeling and dyeing); (B) 3D imaging of optical cleared plant sample, 3D imaging mainly depends on light-sheet fluorescence microscopy, laser scanning confocal microscopy and serial section tomography, after raw images collection of serial optical sections, 3D images of interest target are displayed; (C) Statistical analysis of plant sample after 3D reconstruction, mainly containing histogram statistics of interest parameters, dimension reduction analysis (uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE)) and topological analysis of a multicellular network
| 透明方法 | 主要成分 | 作用 | 优点 | 缺点 | 应用 | 参考文献 |
|---|---|---|---|---|---|---|
| 3DISCO | 四氢呋喃和二氯甲烷 | 脱水和脱脂 | 透明效果好, 荧光蛋白保存良好 | 试剂有毒性, 组织易萎缩 | 鼠脑, 小鼠脊髓、肺和脾, 免疫器官, 肿瘤组织 | Ertu?rk and Bradke, |
| CUBIC | 四乙酸 | 脱色 | 操作简单, 透明效率高, 可重复性强 | 时间较长 | 小鼠组织 | Tainaka et al., |
| CLARITY | 水凝胶 | 组织灌注 | 透明效果好 | 操作复杂, 组织易褐化 | 鼠脑 | Treweek et al., |
| TDE | 硫二甘醇 | RI匹配 | 简单快速, 组织变形、体积变小 | 荧光蛋白易淬灭 | 鼠脑和拟南芥(Arabi- dopsis thaliana)种子 | Staudt et al., |
| 强碱 | 氢氧化钠和氢氧化钾 | 脱脂和除蛋白 | 操作简单, 成本低, 透明效率高 | 组织损伤较大 | 黄花蒿(Artemisia annua)叶片和葡萄(Vitis vinifera)叶片 | 付金娥等, |
| PEA-CLARITY | 细胞壁降解酶 | 水解酶类 | 透明效果好 | 操作复杂, 组织易褐化 | 拟南芥叶片和烟草(Nicotiana tabacum)叶片 | Palmer et al., |
| ClearSee | 木糖醇、脱氧胆酸钠和尿素 | 折射率(RI)匹配 | 保持荧光蛋白稳定性, 减少叶绿体自发荧光 | 时间较长 | 拟南芥幼苗、叶片和雌蕊 | Kurihara et al., |
Table 1 Comparison of clearing techniques in different organisms
| 透明方法 | 主要成分 | 作用 | 优点 | 缺点 | 应用 | 参考文献 |
|---|---|---|---|---|---|---|
| 3DISCO | 四氢呋喃和二氯甲烷 | 脱水和脱脂 | 透明效果好, 荧光蛋白保存良好 | 试剂有毒性, 组织易萎缩 | 鼠脑, 小鼠脊髓、肺和脾, 免疫器官, 肿瘤组织 | Ertu?rk and Bradke, |
| CUBIC | 四乙酸 | 脱色 | 操作简单, 透明效率高, 可重复性强 | 时间较长 | 小鼠组织 | Tainaka et al., |
| CLARITY | 水凝胶 | 组织灌注 | 透明效果好 | 操作复杂, 组织易褐化 | 鼠脑 | Treweek et al., |
| TDE | 硫二甘醇 | RI匹配 | 简单快速, 组织变形、体积变小 | 荧光蛋白易淬灭 | 鼠脑和拟南芥(Arabi- dopsis thaliana)种子 | Staudt et al., |
| 强碱 | 氢氧化钠和氢氧化钾 | 脱脂和除蛋白 | 操作简单, 成本低, 透明效率高 | 组织损伤较大 | 黄花蒿(Artemisia annua)叶片和葡萄(Vitis vinifera)叶片 | 付金娥等, |
| PEA-CLARITY | 细胞壁降解酶 | 水解酶类 | 透明效果好 | 操作复杂, 组织易褐化 | 拟南芥叶片和烟草(Nicotiana tabacum)叶片 | Palmer et al., |
| ClearSee | 木糖醇、脱氧胆酸钠和尿素 | 折射率(RI)匹配 | 保持荧光蛋白稳定性, 减少叶绿体自发荧光 | 时间较长 | 拟南芥幼苗、叶片和雌蕊 | Kurihara et al., |
| [1] | 杜中, 赵世绪, 周海鹰, 王莉莉 (1995). 用微分干涉差显微镜和子房整体透明法观察水稻和小麦的胚囊结构. 农业生物技术学报 3(4), 94-97. |
| [2] | 付金娥, 黎颍菁, 韦树根, 潘丽梅, 胡凤云, 童婷婷 (2017). 叶片气孔保卫细胞周长用于鉴定黄花蒿倍性的研究. 北方园艺 (6), 156-161. |
| [3] | 郭海滨, 卢永根, 冯九焕, 杨秉耀, 刘向东 (2006). 利用激光扫描共聚焦显微术对同源四倍体水稻胚囊形成与发育的进一步观察. 激光生物学报 15, 111-117. |
| [4] | 郝建华, 强胜 (2007). 整体透明技术在植物生物学中的应用实例及其剖析. 植物学通报 24, 490-497. |
| [5] | 李国平, 黄群策, 秦广雍 (2006). 用激光扫描共聚焦显微镜观察雪松花粉和花粉管. 激光生物学报 15, 1-8. |
| [6] | 李亚敏, 薛成志, 李贵叶, 胡章立, 阮双琛, 陈玲玲 (2017). 生物组织光学透明技术研究进展. 生物学杂志 34, 83-88. |
| [7] | 李彦坤, 臧巩固, 赵立宁, 李育君, 唐蜻, 程超华 (2011). 整体透明技术观察悬铃叶苎麻胚胎发育的方法研究. 中国麻业科学 33, 142-146. |
| [8] | 王培新, 张丹, 尚爱加, 侯冰 (2016). 组织透明技术. 神经解剖学杂志 32, 124-128. |
| [9] | 谢兆森, 杜鸿儒, 李建宝, Bondada B (2018). 组织透明法观察葡萄叶片生长过程中气孔与叶脉形态结构特征变化. 植物生理学报 54, 237-246. |
| [10] | 杨弘远 (1988). 植物胚胎学中的整体透明技术. 植物学通报 5, 114-116. |
| [11] |
Aoyagi Y, Kawakami R, Osanai H, Hibi T, Nemoto T (2015). A rapid optical clearing protocol using 2,2'-thiodiethanol for microscopic observation of fixed mouse brain. PLoS One 10, e0116280.
DOI URL |
| [12] |
Bassel GW (2019). Multicellular systems biology: quantifying cellular patterning and function in plant organs using network science. Mol Plant 12, 731-742.
DOI URL |
| [13] |
Becker K, Jährling N, Saghafi S, Weiler R, Dodt HU (2012). Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One 7, e33916.
DOI URL |
| [14] |
Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, Chedotal A (2017). Tridimensional visualization and analysis of early human development. Cell 169, 161-173.
DOI URL |
| [15] |
Berson T, von Wangenheim D, Takáč T, Šamajová O, Rosero A, Ovečka M, Komis G, Stelzer EHK, Šamaj J (2014). Trans-Golgi network localized small GTPase RabA1d is involved in cell plate formation and oscillatory root hair growth. BMC Plant Biol 14, 252.
DOI URL |
| [16] | Cahoon CK, Yu ZL, Wang YF, Guo FL, Unruh JR, Slaughter BD, Hawley RS (2017). Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc Natl Acad Sci USA 114, E6857-E6866. |
| [17] |
Cai RY, Pan CC, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, Bhatia HS, Parra-Damas A, Mrowka L, Theodorou D, Rempfler M, Xavier ALR, Kress BT, Benakis C, Steinke H, Liebscher S, Bechmann I, Liesz A, Menze B, Kerschensteiner M, Nedergaard M, Ertürk A (2019). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat Neurosci 22, 317-327.
DOI URL |
| [18] |
Chung K, Deisseroth K (2013). CLARITY for mapping the nervous system. Nat Methods 10, 508-513.
DOI URL |
| [19] |
Costantini I, Ghobril JP, Di Giovanna AP, Mascaro ALA, Silvestri L, Müllenbroich MC, Onofri L, Conti V, Vanzi F, Sacconi L, Guerrini R, Markram H, Iannello G, Pavone FS (2015). A versatile clearing agent for multi-modal brain imaging. Sci Rep 5, 9808.
DOI PMID |
| [20] | Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, Becker K (2007). Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4, 331-336. |
| [21] |
Duran-Nebreda S, Bassel GW (2017). Bridging scales in plant biology using network science. Trends Plant Sci 22, 1001-1003.
DOI PMID |
| [22] |
Ertürk A, Bradke F (2013). High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO). Exp Neurol 242, 57-64.
DOI URL |
| [23] | Freifeld L, Odstrcil I, Förster D, Ramirez A, Gagnon JA, Randlett O, Costa EK, Asano S, Celiker OT, Gao RX, Martin-Alarcon DA, Reginato P, Dick C, Chen LL, Schoppik D, Engert F, Baier H, Boyden ES (2017). Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc Natl Acad Sci USA 114, E10799-E10808. |
| [24] |
Gardner RO (1975). An overview of botanical clearing technique. Stain Technol 50, 99-105.
PMID |
| [25] |
Gualda E, Moreno N, Tomancak P, Martins GG (2014). Going ‘open' with Mesoscopy: a new dimension on multi- view imaging. Protoplasma 251, 363-372.
DOI URL |
| [26] | Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A (2015). ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518-1529. |
| [27] |
Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A (2011). Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14, 1481-1488.
DOI URL |
| [28] |
Hasegawa J, Sakamoto Y, Nakagami S, Aida M, Sawa S, Matsunaga S (2016). Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol 57, 462-472.
DOI PMID |
| [29] |
Herr JM (1971). A new clearing-squash technique for the study of ovule development in angiosperms. Am J Bot 58, 785-790.
DOI URL |
| [30] |
Horecker BL (1943). The absorption spectra of hemoglobin and its derivatives in the visible and near infra-red regions. J Biol Chem 148, 173-183.
DOI URL |
| [31] | Hou B, Zhang D, Zhao S, Wei MP, Yang ZF, Wang SX, Wang JR, Zhang X, Liu B, Fan LZ, Li Y, Qiu ZL, Zhang C, Jiang TZ (2015). Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat 9, 19. |
| [32] |
Jackson MDB, Xu H, Duran-Nebreda S, Stamm P, Bassel GW (2017). Topological analysis of multicellular complexity in the plant hypocotyl. eLife 6, e26023.
DOI URL |
| [33] |
Jing D, Yi Y, Luo W, Zhang S, Yuan Q, Wang J, Lachika E, Zhao Z, Zhao H (2019). Tissue clearing and its application to bone and dental tissues. J Dent Res 98, 621-631.
DOI PMID |
| [34] |
Jing D, Zhang SW, Luo WJ, Gao XF, Men Y, Ma C, Liu XH, Yi YT, Bugde A, Zhou BO, Zhao ZH, Yuan Q, Feng JQ, Gao L, Ge WP, Zhao H (2018). Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res 28, 803-818.
DOI URL |
| [35] |
Jo Y, Cho H, Park WS, Kim G, Ryu D, Kim YS, Lee M, Park S, Lee MJ, Joo H, Jo H, Lee S, Lee S, Min HS, Heo WD, Park Y (2021). Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning. Nat Cell Biol 23, 1329-1337.
DOI URL |
| [36] |
Johnsen S, Widder EA (1999). The physical basis of transparency in biological tissue: ultrastructure and the minimization of light scattering. J Theor Biol 199, 181-198.
PMID |
| [37] |
Kao P, Nodine MD (2019). Transcriptional activation of Arabidopsis zygotes is required for initial cell divisions. Sci Rep 9, 17159.
DOI URL |
| [38] |
Ke MT, Fujimoto S, Imai T (2013). SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154-1161.
DOI URL |
| [39] |
Ke MT, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, Sato M, Imai T (2016). Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep 14, 2718-2732.
DOI URL |
| [40] |
Kenrick J, Kaul V, Williams EG (1986). Self-incompatibility in Acacia retinodes: site of pollen-tube arrest is the nucellus. Planta 169, 245-250.
DOI PMID |
| [41] |
Kierzkowski D, Routier-Kierzkowska AL (2019). Cellular basis of growth in plants: geometry matters. Curr Opin Plant Biol 47, 56-63.
DOI PMID |
| [42] |
Klingberg A, Hasenberg A, Ludwig-Portugall I, Medyukhina A, Männ L, Brenzel A, Engel DR, Figge MT, Kurts C, Gunzer M (2017). Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J Am Soc Nephrol 28, 452-459.
DOI PMID |
| [43] |
Kubalová I, Černohorská MS, Huranová M, Weisshart K, Houben A, Schubert V (2020). Prospects and limitations of expansion microscopy in chromatin ultrastructure determination. Chromosome Res 28, 355-368.
DOI URL |
| [44] |
Kubota SI, Takahashi K, Nishida J, Morishita Y, Ehata S, Tainaka K, Miyazono K, Ueda HR (2017). Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep 20, 236-250.
DOI URL |
| [45] |
Kurihara D, Mizuta Y, Sato Y, Higashiyama T (2015). ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168-4179.
DOI PMID |
| [46] |
Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C (2013). ClearT: a detergent-and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364-1368.
DOI PMID |
| [47] |
Lucas M, Kenobi K, von Wangenheim D, Voβ U, Swarup K, De Smet I, Van Damme D, Lawrence T, Péret B, Moscardi E, Barbeau D, Godin C, Salt D, Guyomarc'h S, Stelzer EHK, Maizel A, Laplaze L, Bennett MJ (2013). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110, 5229-5234.
DOI URL |
| [48] |
Maizel A, von Wangenheim D, Federici F, Haseloff J, Stelzer EHK (2011). High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J 68, 377-385.
DOI URL |
| [49] |
Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R (1996). Visualizing an olfactory sensory map. Cell 87, 675-686.
PMID |
| [50] |
Montenegro-Johnson TD, Stamm P, Strauss S, Topham AT, Tsagris M, Wood ATA, Smith RS, Bassel GW (2015). Digital single-cell analysis of plant organ development using 3DCellAtlas. Plant Cell 27, 1018-1033.
DOI URL |
| [51] |
Musielak TJ, Slane D, Liebig C, Bayer M (2016). A versatile optical clearing protocol for deep tissue imaging of fluorescent proteins in Arabidopsis thaliana. PLoS One 11, e0161107.
DOI URL |
| [52] | Novák D, Kucharova A, Ovečka M, Komis G, Šamaj J (2016). Developmental nuclear localization and quantification of GFP-tagged EB1c in Arabidopsis root using light-sheet microscopy. Front Plant Sci 6, 1187. |
| [53] |
Ovečka M, Vaškebová L, Komis G, Luptovčiak I, Smertenko A, Šamaj J (2015). Preparation of plants for developmental and cellular imaging by light-sheet microscopy. Nat Protoc 10, 1234-1247.
DOI URL |
| [54] |
Ovečka M, von Wangenheim D, Tomančák P, Šamajová O, Komis G, Šamaj J (2018). Multiscale imaging of plant development by light-sheet fluorescence microscopy. Nat Plants 4, 639-650.
DOI URL |
| [55] |
Palmer WM, Martin AP, Flynn JR, Reed SL, White RG, Furbank RT, Grof CPL (2015). PEA-CLARITY: 3D molecular imaging of whole plant organs. Sci Rep 5, 13492.
DOI URL |
| [56] |
Pan CC, Cai RY, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, Ertürk A (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods 13, 859-867.
DOI |
| [57] |
Petersen CCH (2007). The functional organization of the barrel cortex. Neuron 56, 339-355.
PMID |
| [58] |
Renier N, Wu ZH, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896-910.
DOI URL |
| [59] |
Richardson DS, Lichtman JW (2015). Clarifying tissue clearing. Cell 162, 246-257.
DOI PMID |
| [60] |
Sapala A, Runions A, Smith RS (2019). Mechanics, geometry and genetics of epidermal cell shape regulation: different pieces of the same puzzle. Curr Opin Plant Biol 47, 1-8.
DOI URL |
| [61] |
Seo J, Choe M, Kim SY (2016). Clearing and labeling techniques for large-scale biological tissues. Mol Cells 39, 439-446.
DOI URL |
| [62] |
Shih CY, Peterson CA, Dumbroff EB (1983). A clearing technique for embryos of lipid-storing seeds. Can J Bot 61, 551-555.
DOI URL |
| [63] |
Slattery RA, Grennan AK, Sivaguru M, Sozzani R, Ort DR (2016). Light sheet microscopy reveals more gradual light attenuation in light-green versus dark-green soybean leaves. J Exp Bot 67, 4697-4709.
DOI PMID |
| [64] |
Smith BB (1973). The use of a new clearing technique for the study of early ovule development, megasporogenesis, and megagametogenesis in five species of Cornus L. Am J Bot 60, 322-338.
DOI URL |
| [65] |
Staudt T, Lang MC, Medda R, Engelhardt J, Hell SW (2007). 2,2'-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc Res Tech 70, 1-9.
DOI URL |
| [66] |
Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A, Ueda HR (2015). Advanced CUBIC protocols for whole- brain and whole-body clearing and imaging. Nat Protoc 10, 1709-1727.
DOI PMID |
| [67] |
Susaki EA, Ueda HR (2016). Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem Biol 23, 137-157.
DOI PMID |
| [68] |
Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, Ueda HR (2014). Whole- body imaging with single-cell resolution by tissue decolorization. Cell 159, 911-924.
DOI PMID |
| [69] |
Tainaka K, Kuno A, Kubota SI, Murakami T, Ueda HR (2016). Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu Rev Cell Dev Biol 32, 713-741.
PMID |
| [70] |
Tainaka K, Murakami TC, Susaki EA, Shimizu C, Saito R, Takahashi K, Hayashi-Takagi A, Sekiya H, Arima Y, Nojima S, Ikemura M, Ushiku T, Shimizu Y, Murakami M, Tanaka KF, Iino M, Kasai H, Sasaoka T, Kobayashi K, Miyazono K, Morii E, Isa T, Fukayama M, Kakita A, Ueda HR (2018). Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep 24, 2196-2210.
DOI PMID |
| [71] |
Timmers ACJ (2016). Light microscopy of whole plant organs. J Microsc 263, 165-170.
DOI URL |
| [72] |
Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, Bjorkman PJ, Fowlkes CC, Gradinaru V (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10, 1860-1896.
DOI URL |
| [73] |
Truckenbrodt S, Sommer C, Rizzoli SO, Danzl JG (2019). A practical guide to optimization in X10 expansion microscopy. Nat Protoc 14, 832-863.
DOI PMID |
| [74] |
Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20, 1494-1503.
DOI URL |
| [75] | Tuchin VV (2015). Tissue optics and photonics: light-tissue interaction. J Biomed Photonics Eng 1, 98-134. |
| [76] |
Ueda HR, Ertürk A, Chung K, Gradinaru V, Chédotal A, Tomancak P, Keller PJ (2020). Tissue clearing and its applications in neuroscience. Nat Rev Neurosci 21, 61-79.
DOI URL |
| [77] |
Vergara HM, Pape C, Meechan KI, Zinchenko V, Genoud C, Wanner AA, Mutemi KN, Titze B, Templin RM, Bertucci PY, Simakov O, Dürichen W, Machado P, Savage EL, Schermelleh L, Schwab Y, Friedrich RW, Kreshuk A, Tischer C, Arendt D (2021). Whole-body integration of gene expression and single-cell morphology. Cell 184, 4819-4837.
DOI URL |
| [78] |
Vigouroux RJ, Belle M, Chédotal A (2017). Neuroscience in the third dimension: shedding new light on the brain with tissue clearing. Mol Brain 10, 33.
DOI PMID |
| [79] |
von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EHK, Maizel A (2016). Rules and self- organizing properties of post-embryonic plant organ cell division patterns. Curr Biol 26, 439-449.
DOI PMID |
| [80] | von Wangenheim D, Hauschild R, Friml J (2017). Light sheet fluorescence microscopy of plant roots growing on the surface of a gel. J Vis Exp (119), e55044. |
| [81] |
Vyplelová P, Ovečka M, Šamaj J (2017). Alfalfa root growth rate correlates with progression of microtubules during mitosis and cytokinesis as revealed by environmental light- sheet microscopy. Front Plant Sci 8, 1870.
DOI PMID |
| [82] |
Weissleder R (2001). A clearer vision for in vivo imaging. Nat Biotechnol 19, 316-317.
PMID |
| [83] |
Wilkinson LG, Tucker MR (2017). An optimised clearing protocol for the quantitative assessment of sub-epidermal ovule tissues within whole cereal pistils. Plant Methods 13, 67.
DOI PMID |
| [84] |
Wuyts N, Palauqui JC, Conejero G, Verdeil JL, Granier C, Massonnet C (2010). High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 6, 17.
DOI URL |
| [85] |
Zhang X, Hu ZJ, Guo YY, Shan XY, Li XJ, Lin JX (2020). High-efficiency procedure to characterize, segment, and quantify complex multicellularity in raw micrographs in plants. Plant Methods 16, 100.
DOI PMID |
| [86] |
Zhang X, Man Y, Zhuang XH, Shen JB, Zhang Y, Cui YN, Yu M, Xing JJ, Wang GC, Lian N, Hu ZJ, Ma LY, Shen WW, Yang SY, Xu HM, Bian JH, Jing YP, Li XJ, Li RL, Mao TL, Jiao YL, Sodmergen, Ren HY, Lin JX (2021). Plant multiscale networks: charting plant connectivity by multi-level analysis and imaging techniques. Sci China Life Sci 64, 1392-1422.
DOI URL |
| [87] |
Zhu MY, Roeder AH (2020). Plants are better engineers: the complexity of plant organ morphogenesis. Curr Opin Genet Dev 63, 16-23.
DOI URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||