

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (2): 214-232.DOI: 10.11983/CBB22220 cstr: 32102.14.CBB22220
• INVITED REVIEWS • Previous Articles Next Articles
Yubin Xiao1,2, Zixu Zhang1, Yuzhu Wang1, Huan Liu2,3,*( ), Letian Chen1,*(
), Letian Chen1,*( )
)
Received:2022-09-12
Accepted:2023-01-10
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: Yubin Xiao, Zixu Zhang, Yuzhu Wang, Huan Liu, Letian Chen. Research Progress of Spatiotemporal Transcriptomes[J]. Chinese Bulletin of Botany, 2023, 58(2): 214-232.
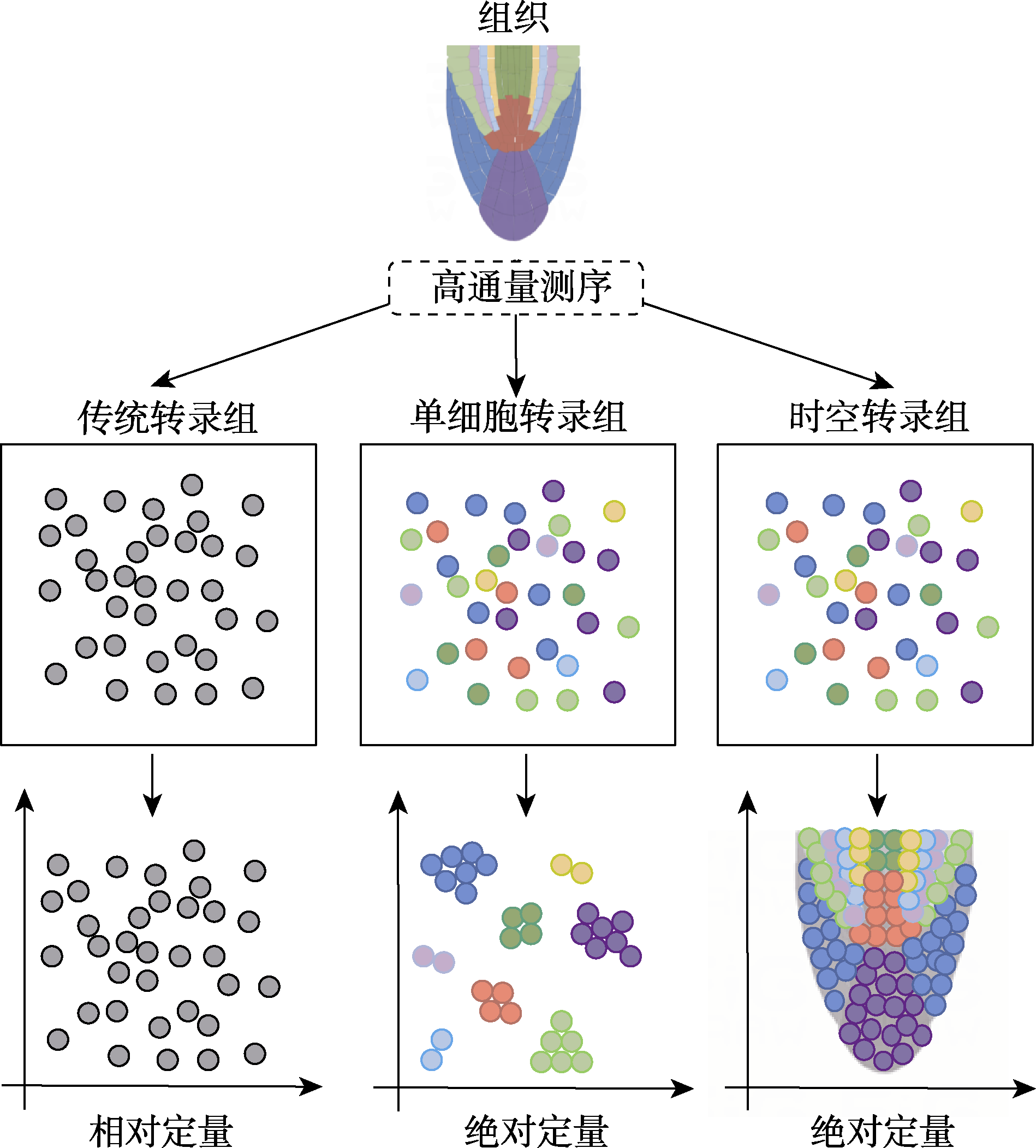
Figure 1 Comparison of transcriptomic technologies The circles with different colors represent different cell types, while the gray circles represent that RNA-seq cannot distinguish different cell types.
| 动物 | 植物 | |
|---|---|---|
| 特有结构 | ? | 细胞壁(初生壁和次生壁)、液泡和叶绿体 |
| 包埋方法 | 冷冻包埋和石蜡包埋 | 冷冻包埋 |
| 组织切片 | 相对简单 | 相对困难, 高度木质化组织容易开裂、液泡易产生结晶等 |
| 染色方法 | 苏木精-伊红染色和ssDNA染色 | 甲苯铵蓝染色、ssDNA染色和钙白染色 |
| 组织透化 | 透化细胞膜 | 透化细胞壁和细胞膜, 且不同植物细胞的细胞壁成分存在较大异质性 |
| 组织移除 | 相对简单 | 高度成熟组织的降解可能需要较长的时间和苛刻的组织移除操作, 可能会影响逆转录产物 |
| 参考基因组 | 参考基因组较为完善 | 参考基因组有待进一步完善 |
| 技术平台 | 大量时空组技术优先适配于动物组织 | 时空组技术在植物组织中的应用条件仍有待进一步优化 |
| 时空组学文献数量 | ~5000 | <1000 |
Table 1 Comparison of spatiotemporal transcriptomes in animals and plants
| 动物 | 植物 | |
|---|---|---|
| 特有结构 | ? | 细胞壁(初生壁和次生壁)、液泡和叶绿体 |
| 包埋方法 | 冷冻包埋和石蜡包埋 | 冷冻包埋 |
| 组织切片 | 相对简单 | 相对困难, 高度木质化组织容易开裂、液泡易产生结晶等 |
| 染色方法 | 苏木精-伊红染色和ssDNA染色 | 甲苯铵蓝染色、ssDNA染色和钙白染色 |
| 组织透化 | 透化细胞膜 | 透化细胞壁和细胞膜, 且不同植物细胞的细胞壁成分存在较大异质性 |
| 组织移除 | 相对简单 | 高度成熟组织的降解可能需要较长的时间和苛刻的组织移除操作, 可能会影响逆转录产物 |
| 参考基因组 | 参考基因组较为完善 | 参考基因组有待进一步完善 |
| 技术平台 | 大量时空组技术优先适配于动物组织 | 时空组技术在植物组织中的应用条件仍有待进一步优化 |
| 时空组学文献数量 | ~5000 | <1000 |
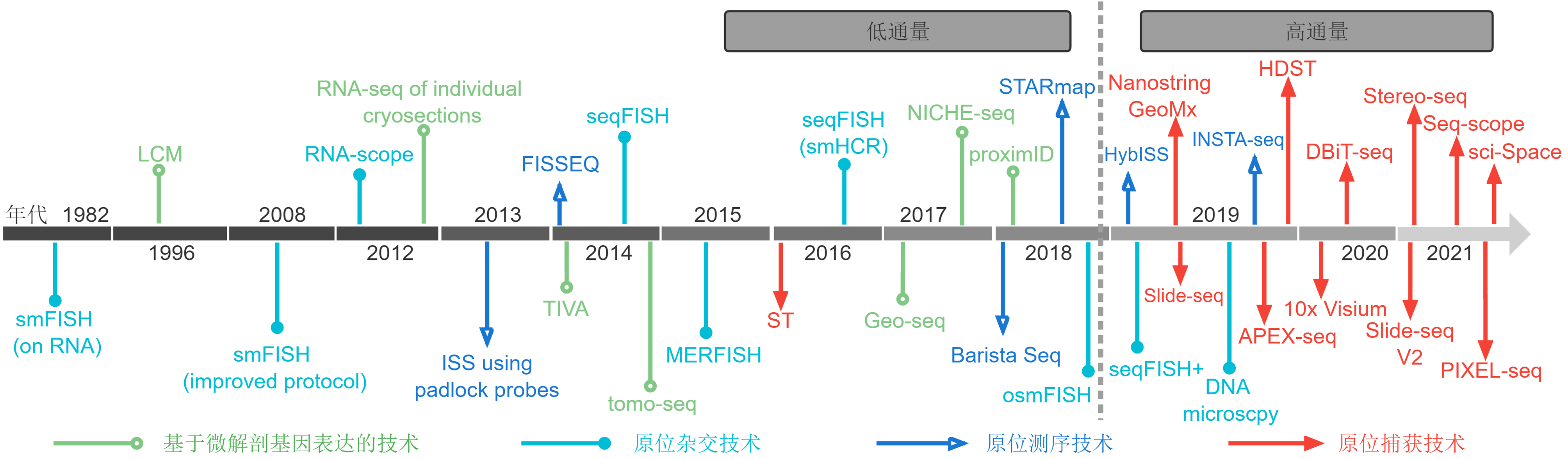
Figure 2 The evolution of spatiotemporal transcriptomes Different colors and shapes represent different types of spatiotemporal transcriptomic techniques. The end of 2018 is a watershed of spatiotemporal transcriptomic techniques: before 2018, the techniques were low throughput, while those after 2018 are high throughput. smFISH: Single-molecule fluorescence in situ hybridization; LCM: Laser-capture microdissection; ISS: In situ sequencing; FISSEQ: Fluorescent in situ sequencing; TIVA: Transcriptome in vivo analysis; seqFISH: Sequential fluorescence in situ hybridization; tomo-seq: RNA tomography; MERFISH: Multiplexed error-robust fluorescence in situ hybridization; ST: Spatial transcriptomics; Geo-seq: Geographical position sequencing; BaristaSeq: Barcode in situ targeted sequencing; STARmap: Spatially-resolved transcript amplicon readout mapping; osmFISH: Ouroboros single-molecule fluorescence in situ hybridization; HybISS: Hybridization-based in situ sequencing; INSTA-seq: In situ transcriptome accessibility sequencing; DBiT-seq: Deterministic barcoding in tissue for spatial omics sequencing; PIXEL-seq: Polony-indexed library-sequencing
| 转录组技术 | 空间分辨率 | 细胞通量 | 优缺点 | 应用范围 | 参考文献 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 传统转录组学 | 组织水平 | 高 | + 成本低, 检测速度快 ? 掩盖了细胞的时空异质性 | 动植物 微生物 | Costa et al., | |||||
| 单细胞转录组学 | 单细胞水平 | 高 | + 通量高, 检测深度较深 ? 忽略了细胞的空间信息 | 动植物 微生物 | Kolodziejczyk et al., | |||||
| 时空转录组学 | 基于微解剖基因表达的技术 | LCM | 单细胞水平 | 低 | + 可人工挑选感兴趣的细胞 ? 通量低, 操作难度大 | 动植物 微生物 | Emmert-buck et al., | |||
| TIVA | 单细胞水平 | 低 | + 能应用于活细胞 ? 通量低, 分析结果有限 | 动物 | Lovatt et al., | |||||
| Tomo-seq | 单个切片 | 低 | + 可构建3D轮廓表达谱 ? 需多个相同的生物样本, 不能应用于人体样本 | 动物 | Junker et al., | |||||
| Geo-seq | 单细胞水平 | 低 | + 比LCM更灵敏 ? 通量低 | 动物 | Chen et al., | |||||
| NICHE-seq | 单细胞水平 | 高 | + 通量高 ? 只能确定特定生态位的空间位置, 不能应用于人体样本 | 动物 | Medaglia et al., | |||||
| proximID | 单细胞水平 | 低 | + 保留部分细胞相互作用的互作结构 ? 需基于LCM进行细胞分离, 通量低 | 动物 | Boisset et al., | |||||
| 原位杂交技术 | smFISH | 亚细胞水平 | 低 | + 灵敏度高, 检测效率相对较高 ? 检测的RNA数量有限 | 动植物 微生物 | Femino et al., | ||||
| RNA-scope | 亚细胞水平 | 低 | + 特异性高, 信噪比极高, 灵敏度高 ? 通量低 | 动植物 微生物 | Wang et al., | |||||
| seqFISH | 亚细胞水平 | 低 | + 高度多重化 ? 需要专门的设备, 视野受限 | 动物 | Lubeck et al., | |||||
| MERFISH | 单细胞水平 | 较高 | + 高度多重化 ? 需要专门的设备, 视野受限 | 动物 | Chen et al., | |||||
| osmFISH | 亚细胞水平 | 较高 | + 可检测更大的组织区域, 半自动化 ? 通量相对较低 | 动物 | Codeluppi et al., | |||||
| seqFISH+ | 单细胞水平 | 较高 | + 极度多重化 ? 需要专门的设备, 视野受限 | 动物 | Eng et al., | |||||
| DNA microscpy | 单细胞水平 | 较高 | + 不依赖光或任何类型的光学器件进行组织的基因表达成像 ? 仅在乳腺癌细胞中应用过 | 动物 | Weinstein et al., | |||||
| 原位测序技术 | ISS using padlock probes | 亚细胞水平 | 较低 | + 亚细胞分辨率检测SNV的能力 ? 转录本检测灵敏度低 | 动植物 微生物 | Ke et al., | ||||
| FISSEQ | 亚细胞水平 | 较低 | + 可捕获所有类型的RNA ? 转录本检测灵敏度相对较低 | 动物 | Lee et al., | |||||
| Barista Seq | 亚细胞水平 | 较低 | + 探针缺口内读取长度可达15个碱基 ? 仅在培养细胞上应用过 | 动物 | Chen et al., | |||||
| STARmap | 亚细胞水平 | 较高 | + 不需要反转录步骤, 灵敏度高 ? 转录本检测灵敏度低, 视野有限 | 动物 | Mano et al., 2019 | |||||
| INSTA-seq | 亚细胞水平 | 较高 | + 一种非靶标的方法, 原位测序区域条形码, 异位测序转录本 ? 检测灵敏度相对较低 | 动物 | Fürth et al., | |||||
| 原位捕获技术 | ST/10× Visium | 100 μm/ 55 μm | 高 | + 可检测完整组织切片中总mRNA ? 条形码区域可能覆盖多个细胞, 未达到单细胞水平 | 动植物 微生物 | St?hl et al., | ||||
| Nanostring GeoMx | 10 μm | 高 | + 在蛋白质/RNA分析之间选择高度自动化 ? 使用较小ROI时灵敏度低, 需要手动选择区域 | 动物 微生物 | Merritt et al., | |||||
| Slide-seq | 10 μm | 高 | + 高分辨率 ? 检测灵敏度相对较低 | 动物 | Rodriques et al., | |||||
| 时空转录组学 | 原位捕获技术 | APEX-seq | 亚细胞水平 | 高 | + 可以在活细胞中进行 ? 需要让特定APEX2酶在特定的亚细胞区域重组表达, 并不适用于正常组织 | 动物 | Fazal et al., | |||
| HDST | 2 μm | 高 | + 高分辨率 ? 稀疏数据需要分箱, 检测灵敏度相对较低 | 动物 | Vickovic et al., | |||||
| DBiT-seq | 10 μm | 高 | + 可同时构建mRNA和蛋白质的空间多组学图谱 ? 检测灵敏度相对较低 | 动物 | Liu et al., | |||||
| Stereo-seq | 0.5 μm | 高 | + 亚细胞分辨率和厘米级视野 ? 检测灵敏度相对较低 | 动植物 微生物 | Chen et al., | |||||
| Seq-scope | 0.5 μm | 高 | + 亚细胞分辨率 ? 检测灵敏度相对较低 | 动物 | Cho et al., | |||||
| PIXEL-seq | 1 μm | 高 | + 亚细胞分辨率, 成本低 ? 检测灵敏度相对较低 | 动物 | Fu et al., | |||||
| sci-Space | 单细胞水平 | 高 | + 利用sci-RNA-seq进行测序, 检测灵敏度相对较高 ? 依赖于sci-Plex进行空间信息记录, 分辨率相对较低, 只进行核基因测序 | 动物 | Srivatsan et al., | |||||
Table 2 Comparison of different transcriptomic techniques
| 转录组技术 | 空间分辨率 | 细胞通量 | 优缺点 | 应用范围 | 参考文献 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 传统转录组学 | 组织水平 | 高 | + 成本低, 检测速度快 ? 掩盖了细胞的时空异质性 | 动植物 微生物 | Costa et al., | |||||
| 单细胞转录组学 | 单细胞水平 | 高 | + 通量高, 检测深度较深 ? 忽略了细胞的空间信息 | 动植物 微生物 | Kolodziejczyk et al., | |||||
| 时空转录组学 | 基于微解剖基因表达的技术 | LCM | 单细胞水平 | 低 | + 可人工挑选感兴趣的细胞 ? 通量低, 操作难度大 | 动植物 微生物 | Emmert-buck et al., | |||
| TIVA | 单细胞水平 | 低 | + 能应用于活细胞 ? 通量低, 分析结果有限 | 动物 | Lovatt et al., | |||||
| Tomo-seq | 单个切片 | 低 | + 可构建3D轮廓表达谱 ? 需多个相同的生物样本, 不能应用于人体样本 | 动物 | Junker et al., | |||||
| Geo-seq | 单细胞水平 | 低 | + 比LCM更灵敏 ? 通量低 | 动物 | Chen et al., | |||||
| NICHE-seq | 单细胞水平 | 高 | + 通量高 ? 只能确定特定生态位的空间位置, 不能应用于人体样本 | 动物 | Medaglia et al., | |||||
| proximID | 单细胞水平 | 低 | + 保留部分细胞相互作用的互作结构 ? 需基于LCM进行细胞分离, 通量低 | 动物 | Boisset et al., | |||||
| 原位杂交技术 | smFISH | 亚细胞水平 | 低 | + 灵敏度高, 检测效率相对较高 ? 检测的RNA数量有限 | 动植物 微生物 | Femino et al., | ||||
| RNA-scope | 亚细胞水平 | 低 | + 特异性高, 信噪比极高, 灵敏度高 ? 通量低 | 动植物 微生物 | Wang et al., | |||||
| seqFISH | 亚细胞水平 | 低 | + 高度多重化 ? 需要专门的设备, 视野受限 | 动物 | Lubeck et al., | |||||
| MERFISH | 单细胞水平 | 较高 | + 高度多重化 ? 需要专门的设备, 视野受限 | 动物 | Chen et al., | |||||
| osmFISH | 亚细胞水平 | 较高 | + 可检测更大的组织区域, 半自动化 ? 通量相对较低 | 动物 | Codeluppi et al., | |||||
| seqFISH+ | 单细胞水平 | 较高 | + 极度多重化 ? 需要专门的设备, 视野受限 | 动物 | Eng et al., | |||||
| DNA microscpy | 单细胞水平 | 较高 | + 不依赖光或任何类型的光学器件进行组织的基因表达成像 ? 仅在乳腺癌细胞中应用过 | 动物 | Weinstein et al., | |||||
| 原位测序技术 | ISS using padlock probes | 亚细胞水平 | 较低 | + 亚细胞分辨率检测SNV的能力 ? 转录本检测灵敏度低 | 动植物 微生物 | Ke et al., | ||||
| FISSEQ | 亚细胞水平 | 较低 | + 可捕获所有类型的RNA ? 转录本检测灵敏度相对较低 | 动物 | Lee et al., | |||||
| Barista Seq | 亚细胞水平 | 较低 | + 探针缺口内读取长度可达15个碱基 ? 仅在培养细胞上应用过 | 动物 | Chen et al., | |||||
| STARmap | 亚细胞水平 | 较高 | + 不需要反转录步骤, 灵敏度高 ? 转录本检测灵敏度低, 视野有限 | 动物 | Mano et al., 2019 | |||||
| INSTA-seq | 亚细胞水平 | 较高 | + 一种非靶标的方法, 原位测序区域条形码, 异位测序转录本 ? 检测灵敏度相对较低 | 动物 | Fürth et al., | |||||
| 原位捕获技术 | ST/10× Visium | 100 μm/ 55 μm | 高 | + 可检测完整组织切片中总mRNA ? 条形码区域可能覆盖多个细胞, 未达到单细胞水平 | 动植物 微生物 | St?hl et al., | ||||
| Nanostring GeoMx | 10 μm | 高 | + 在蛋白质/RNA分析之间选择高度自动化 ? 使用较小ROI时灵敏度低, 需要手动选择区域 | 动物 微生物 | Merritt et al., | |||||
| Slide-seq | 10 μm | 高 | + 高分辨率 ? 检测灵敏度相对较低 | 动物 | Rodriques et al., | |||||
| 时空转录组学 | 原位捕获技术 | APEX-seq | 亚细胞水平 | 高 | + 可以在活细胞中进行 ? 需要让特定APEX2酶在特定的亚细胞区域重组表达, 并不适用于正常组织 | 动物 | Fazal et al., | |||
| HDST | 2 μm | 高 | + 高分辨率 ? 稀疏数据需要分箱, 检测灵敏度相对较低 | 动物 | Vickovic et al., | |||||
| DBiT-seq | 10 μm | 高 | + 可同时构建mRNA和蛋白质的空间多组学图谱 ? 检测灵敏度相对较低 | 动物 | Liu et al., | |||||
| Stereo-seq | 0.5 μm | 高 | + 亚细胞分辨率和厘米级视野 ? 检测灵敏度相对较低 | 动植物 微生物 | Chen et al., | |||||
| Seq-scope | 0.5 μm | 高 | + 亚细胞分辨率 ? 检测灵敏度相对较低 | 动物 | Cho et al., | |||||
| PIXEL-seq | 1 μm | 高 | + 亚细胞分辨率, 成本低 ? 检测灵敏度相对较低 | 动物 | Fu et al., | |||||
| sci-Space | 单细胞水平 | 高 | + 利用sci-RNA-seq进行测序, 检测灵敏度相对较高 ? 依赖于sci-Plex进行空间信息记录, 分辨率相对较低, 只进行核基因测序 | 动物 | Srivatsan et al., | |||||
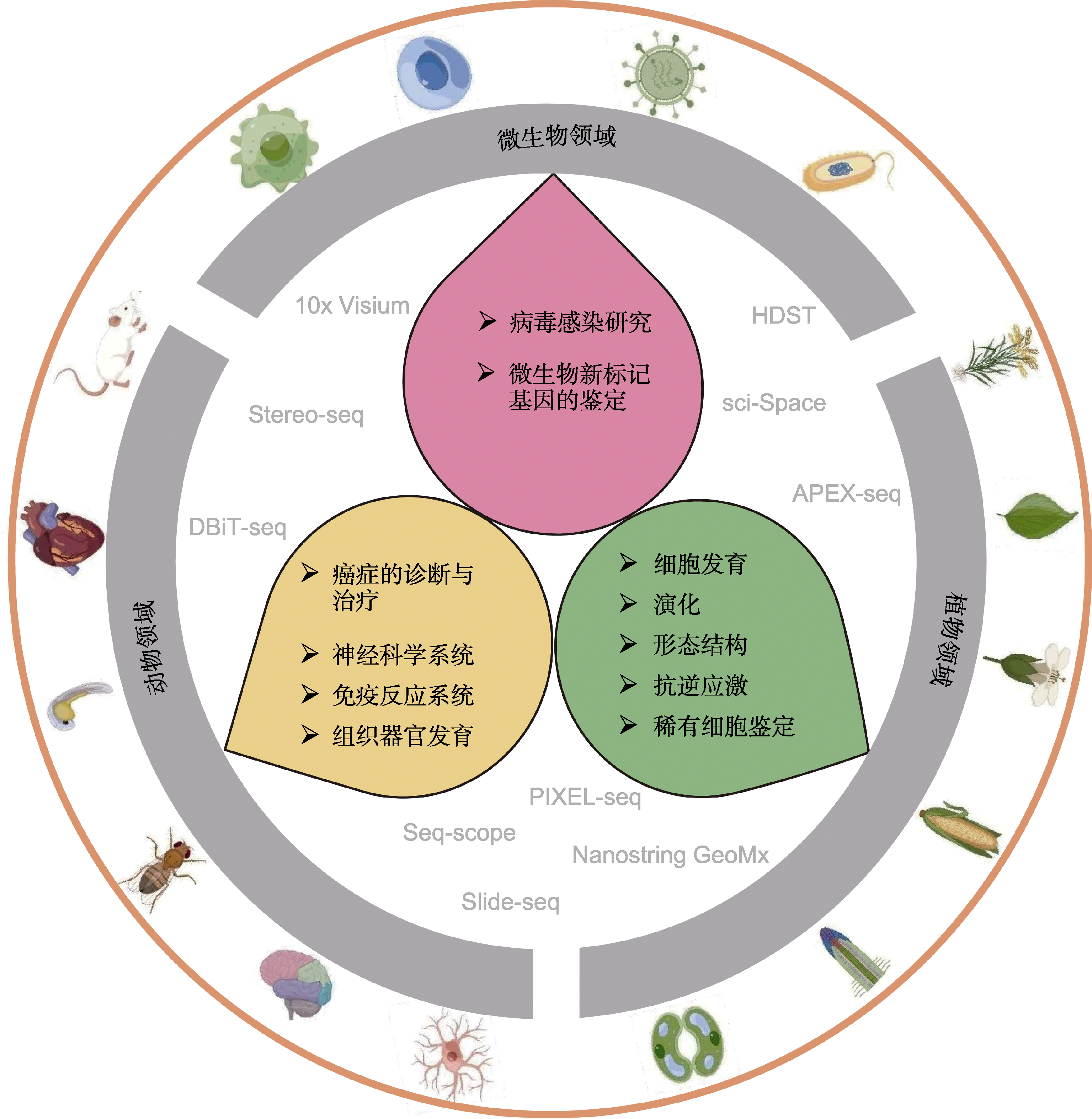
Figure 3 The major research materials and application fields for spatiotemporal transcriptomics in animals, plants and microorganisms Yellow, green and pink backgrounds represent the major application fields of spatiotemporal transcriptomics in animals, plants and microorganisms, respectively.
| [1] | 操利超, 巴颖, 张核子 (2022). 单细胞测序方法研究进展. 生物信息学 20, 91-99. |
| [2] | 崔桂忠, 彭广敦, 景乃禾 (2020). 高精度时空转录组揭示小鼠早期胚胎三胚层细胞谱系发生过程. 中国细胞生物学学报 42, 1-8. |
| [3] |
牛艳丽, 柏胜龙, 王麒云, 刘凌云 (2017). 单细胞组学技术及其在植物保卫细胞研究中的应用. 植物学报 52, 788-796.
DOI |
| [4] | 邱丹丹, 蒋松 (2022). 空间转录组技术在肾脏疾病研究中的应用. 肾脏病与透析肾移植杂志 31, 256-260. |
| [5] | 项铮, 苏存锦, 潘杰 (2022). 空间转录物组学技术进展及其在神经科学领域中的应用. 中国生物化学与分子生物学报 38, 1486-1492. |
| [6] | 杨佳凤, 陈鹏璐, 龚熹 (2021). 单细胞转录组测序技术在细胞分类中的应用. 中国细胞生物学学报 43, 476-483. |
| [7] | 赵宇豪, 李永盛, 央茂, 王许安, 吴文广, 刘颖斌 (2022). 空间转录组测序技术在肿瘤发生发展机制中的应用及前景. 中华医学杂志 102, 1551-1554. |
| [8] | 朱彬彬, 刘亚慧, 齐大屯, 程倩倩, 李高寒, 高传玉 (2022). 单细胞测序和空间转录组学在心血管疾病研究中的进展. 中国心血管病研究 20, 497-502. |
| [9] |
Andersson A, Larsson L, Stenbeck L, Salmén F, Ehinger A, Wu SZ, Al-Eryani G, Roden D, Swarbrick A, Borg Å, Frisén J, Engblom C, Lundeberg J (2021). Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat Commun 12, 6012.
DOI PMID |
| [10] |
Asp M, Bergenstråhle J, Lundeberg J (2020). Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays 42, 1900221.
DOI URL |
| [11] |
Asp M, Giacomello S, Larsson L, Wu CL, Fürth D, Qian XY, Wärdell E, Custodio J, Reimegård J, Salmén F, Österholm C, Ståhl PL, Sundström E, Åkesson E, Bergmann O, Bienko M, Månsson-Broberg A, Nilsson M, Sylvén C, Lundeberg J (2019). A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179, 1647-1660.
DOI PMID |
| [12] |
Baccin C, Al-Sabah J, Velten L, Helbling PM, Grünschläger F, Hernández-Malmierca P, Nombela-Arrieta C, Steinmetz LM, Trumpp A, Haas S (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol 22, 38-48.
DOI PMID |
| [13] |
Boisset JC, Vivié J, Grün D, Muraro MJ, Lyubimova A, van Oudenaarden A (2018). Mapping the physical network of cellular interactions. Nat Methods 15, 547-553.
DOI |
| [14] |
Boyd DF, Allen EK, Randolph AG, Guo XZJ, Weng YC, Sanders CJ, Bajracharya R, Lee NK, Guy CS, Vogel P, Guan WD, Li YM, Liu XQ, Novak T, Newhams MM, Fabrizio TP, Wohlgemuth N, Mourani PM, PALISI Pediatric Intensive Care Influenza PICFLU Investigators, Wight TN, Schultz-Cherry S, Cormier SA, Shaw-Saliba K, Pekosz A, Rothman RE, Chen KF, Yang ZF, Webby RJ, Zhong NS, Crawford JC, Thomas PG (2020). Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature 587, 466-471.
DOI |
| [15] |
Butler D, Mozsary C, Meydan C, Foox J, Rosiene J, Shaiber A, Danko D, Afshinnekoo E, MacKay M, Sedlazeck FJ, Ivanov NA, Sierra M, Pohle D, Zietz M, Gisladottir U, Ramlall V, Sholle ET, Schenck EJ, Westover CD, Hassan C, Ryon K, Young B, Bhattacharya C, Ng DL, Granados AC, Santos YA, Servellita V, Federman S, Ruggiero P, Fungtammasan A, Chin CS, Pearson NM, Langhorst BW, Tanner NA, Kim Y, Reeves JW, Hether TD, Warren SE, Bailey M, Gawrys J, Meleshko D, Xu D, Couto-Rodriguez M, Nagy-Szakal D, Barrows J, Wells H, O'Hara NB, Rosenfeld JA, Chen Y, Steel PAD, Shemesh AJ, Xiang J, Thierry-Mieg J, Thierry- Mieg D, Iftner A, Bezdan D, Sanchez E, Campion TR Jr, Sipley J, Cong L, Craney A, Velu P, Melnick AM, Shapira S, Hajirasouliha I, Borczuk A, Iftner T, Salvatore M, Loda M, Westblade LF, Cushing M, Wu SX, Levy S, Chiu C, Schwartz RE, Tatonetti N, Rennert H, Imielinski M, Mason CE (2021). Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat Commun 12, 1660.
DOI PMID |
| [16] |
Cao JY, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu XJ, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661-667.
DOI PMID |
| [17] |
Carlberg K, Korotkova M, Larsson L, Catrina AI, Ståhl PL, Malmström V (2019). Exploring inflammatory signatures in arthritic joint biopsies with spatial transcriptomics. Sci Rep 9, 18975.
DOI PMID |
| [18] |
Chen A, Liao S, Cheng MN, Ma KL, Wu L, Lai YW, Qiu XJ, Yang J, Xu JS, Hao SJ, Wang X, Lu HF, Chen X, Liu X, Huang X, Li Z, Hong Y, Jiang YJ, Peng J, Liu S, Shen MZ, Liu CY, Li QS, Yuan Y, Wei XY, Zheng HW, Feng WM, Wang ZF, Liu Y, Wang ZH, Yang YZ, Xiang HT, Han L, Qin BM, Guo PC, Lai GY, Muñoz-Cánoves P, Maxwell PH, Thiery JP, Wu QF, Zhao FX, Chen BC, Li M, Dai X, Wang S, Kuang HY, Hui JH, Wang LQ, Fei JF, Wang O, Wei XF, Lu HR, Wang B, Liu SP, Gu Y, Ni M, Zhang WW, Mu F, Yin Y, Yang HM, Lisby M, Cornall RJ, Mulder J, Uhlén M, Esteban MA, Li YX, Liu LQ, Xu X, Wang J (2022). Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777-1792.
DOI PMID |
| [19] |
Chen J, Suo SB, Tam PPL, Han JDJ, Peng GD, Jing NH (2017). Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat Protoc 12, 566-580.
DOI PMID |
| [20] | Chen KH, Boettiger AN, Moffitt JR, Wang SY, Zhuang XW (2015). Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. |
| [21] | Chen XY, Sun YC, Church GM, Lee JH, Zador AM (2018). Efficient in situ barcode sequencing using padlock probe- based BaristaSeq. Nucleic Acids Res 46, e22. |
| [22] |
Cho CS, Xi JY, Si YC, Park SR, Hsu JE, Kim M, Jun G, Kang HM, Lee JH (2021). Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559-3572.
DOI URL |
| [23] |
Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, Linnarsson S (2018). Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods 15, 932-935.
DOI PMID |
| [24] | Combs PA, Eisen MB (2013). Sequencing mRNA from cryo-sliced Drosophila embryos to determine genome-wide spatial patterns of gene expression. PLoS One 8, e71820. |
| [25] | Costa V, Angelini C, De Feis I, Ciccodicola A (2010). Uncovering the complexity of transcriptomes with RNA- Seq. J Biomed Biotechnol 2010, 853916. |
| [26] |
Downes DJ, Cross AR, Hua P, Roberts N, Schwessinger R, Cutler AJ, Munis AM, Brown J, Mielczarek O, de Andrea CE, Melero I, COvid-19 Multi-Omics Blood ATlas COMBAT Consortium, Gill DR, Hyde SC, Knight JC, Todd JA, Sansom SN, Issa F, Davies JOJ, Hughes JR (2021). Identification of LZTFL1as a candidate effector gene at a COVID-19 risk locus. Nat Genet 53, 1606-1615.
DOI |
| [27] |
Eisenstein M (2022). Seven technologies to watch in 2022. Nature 601, 658-661.
DOI |
| [28] |
Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang ZP, Goldstein SR, Weiss RA, Liotta LA (1996). Laser capture microdissection. Science 274, 998-1001.
DOI PMID |
| [29] |
Eng CHL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun JN, Cronin C, Karp C, Yuan GC, Cai L (2019). Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235-239.
DOI |
| [30] |
Fawkner-Corbett D, Antanaviciute A, Parikh K, Jagielowicz M, Gerós AS, Gupta T, Ashley N, Khamis D, Fowler D, Morrissey E, Cunningham C, Johnson PRV, Koohy H, Simmons A (2021). Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 184, 810-826.
DOI PMID |
| [31] |
Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY (2019). Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473-490.
DOI PMID |
| [32] |
Femino AM, Fay FS, Fogarty K, Singer RH (1998). Visualization of single RNA transcripts in situ. Science 280, 585-590.
PMID |
| [33] |
Fu XN, Sun L, Chen JY, Dong RZ, Lin Y, Palmiter RD, Lin S, Gu LC (2021). Continuous polony gels for tissue mapping with high resolution and RNA capture efficiency. BioRxiv doi: 10.1101/2021.03.17.435795.
DOI |
| [34] |
Fu XN, Sun L, Dong RZ, Chen JY, Silakit R, Condon LF, Lin Y, Lin S, Palmiter RD, Gu LC (2022). Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain. Cell 185, 4621-4633.
DOI PMID |
| [35] |
Fürth D, Hatini V, Lee JH (2019). In situ transcriptome accessibility sequencing (INSTA-seq). BioRxiv doi: 10 1101/ 722819.
DOI |
| [36] |
Gao SW, Shi Q, Zhang YF, Liang GX, Kang ZX, Huang BF, Ma DY, Wang L, Jiao JW, Fang XD, Xu CR, Liu LQ, Xu X, Göttgens B, Li C, Liu F (2022). Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res 32, 38-53.
DOI |
| [37] |
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26, 317-325.
DOI PMID |
| [38] |
Giacomello S, Lundeberg J (2018). Preparation of plant tissue to enable spatial transcriptomics profiling using barcoded microarrays. Nat Protoc 13, 2425-2446.
DOI PMID |
| [39] |
Giacomello S, Salmén F, Terebieniec BK, Vickovic S, Navarro JF, Alexeyenko A, Reimegård J, McKee LS, Mannapperuma C, Bulone V, Ståhl PL, Sundström JF, Street NR, Lundeberg J (2017). Spatially resolved transcriptome profiling in model plant species. Nat Plants 3, 17061.
DOI PMID |
| [40] |
Giolai M, Verweij W, Lister A, Heavens D, Macaulay I, Clark MD (2019). Spatially resolved transcriptomics reveals plant host responses to pathogens. Plant Methods 15, 114.
DOI PMID |
| [41] |
Gough A, Stern AM, Maier J, Lezon T, Shun TY, Chennubhotla C, Schurdak ME, Haney SA, Taylor DL (2017). Biologically relevant heterogeneity: metrics and practical insights. SLAS Discov 22, 213-237.
DOI PMID |
| [42] |
Gurazada SGR, Cox KL Jr, Czymmek KJ, Meyers BC (2021). Space: the final frontier-achieving single-cell, spatially resolved transcriptomics in plants. Emerg Top Life Sci 5, 179-188.
DOI PMID |
| [43] |
Hou XL, Yang Y, Li P, Zeng ZP, Hu WL, Zhe RL, Liu XQ, Tang DE, Ou ML, Dai Y (2021). Integrating spatial transcriptomics and single-cell RNA-seq reveals the gene expression profling of the human embryonic liver. Front Cell Dev Biol 9, 652408.
DOI URL |
| [44] |
Ji AL, Rubin AJ, Thrane K, Jiang SZ, Reynolds DL, Meyers RM, Guo MG, George BM, Mollbrink A, Bergenstråhle J, Larsson L, Bai YH, Zhu BK, Bhaduri A, Meyers JM, Rovira-Clavé X, Hollmig ST, Aasi SZ, Nolan GP, Lundeberg J, Khavari PA (2020). Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497-514.
DOI PMID |
| [45] |
Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A (2014). Genome-wide RNA tomography in the zebrafish embryo. Cell 159, 662-675.
DOI PMID |
| [46] | Ke RQ, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M (2013). In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 10, 857-860. |
| [47] |
Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA (2015). The technology and biology of single-cell RNA sequencing. Mol Cell 58, 610-620.
DOI PMID |
| [48] |
Kulasinghe A, Tan CW, dos Santos Miggiolaro AFR, Monkman J, SadeghiRad H, Bhuva DD, da Silva Motta Junior J, Vaz de Paula CB, Nagashima S, Baena CP, Souza-Fonseca-Guimaraes P, de Noronha L, McCulloch T, Rossi GR, Cooper C, Tang B, Short KR, Davis MJ, Souza-Fonseca-Guimaraes F, Belz GT, O'Byrne K (2022). Profiling of lung SARS-CoV-2 and influenza virus infection dissects virus-specific host responses and gene signatures. Eur Respir J 59, 2101881.
DOI URL |
| [49] |
Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, Peters DT, Turczyk BM, Marblestone AH, Inverso SA, Bernard A, Mali P, Rios X, Aach J, Church GM (2014). Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360-1363.
DOI URL |
| [50] |
Lee JK, Wang JG, Sa JK, Ladewig E, Lee HO, Lee IH, Kang HJ, Rosenbloom DS, Camara PG, Liu ZQ, van Nieuwenhuizen P, Jung SW, Choi SW, Kim J, Chen A, Kim KT, Shin S, Seo YJ, Oh JM, Shin YJ, Park CK, Kong DS, Seol HJ, Blumberg A, Lee JI, Iavarone A, Park WY, Rabadan R, Nam DH (2017). Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 49, 594-599.
DOI |
| [51] |
Lein E, Borm LE, Linnarsson S (2017). The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64-69.
DOI PMID |
| [52] |
Liao J, Lu XY, Shao X, Zhu L, Fan XH (2021). Uncovering an organ's molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol 39, 43-58.
DOI PMID |
| [53] | Lieben L (2017). Spatial transcriptomics in plants. Nat Rev Genet 18, 394. |
| [54] | Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523-536. |
| [55] |
Liu C, Li R, Li Y, Lin XM, Zhao KC, Liu Q, Wang SW, Yang XQ, Shi XY, Ma YT, Pei CY, Wang H, Bao WD, Hui JH, Yang T, Xu ZC, Lai TT, Berberoglu MA, Sahu SK, Esteban MA, Ma KL, Fan GY, Li YX, Liu SP, Chen A, Xu X, Dong ZQ, Liu LQ (2022a). Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis. Dev Cell 57, 1284-1298.
DOI URL |
| [56] |
Liu Y, Yang MY, Deng YX, Su G, Enninful A, Guo CC, Tebaldi T, Zhang D, Kim D, Bai ZL, Norris E, Pan A, Li JT, Xiao Y, Halene S, Fan R (2020). High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665-1681.
DOI PMID |
| [57] |
Liu YY, Li CH, Han Y, Li RC, Cui F, Zhang H, Su XS, Liu XW, Xu GX, Wan SB, Li GW (2022b). Spatial transcriptome analysis on peanut tissues shed light on cell heterogeneity of the peg. Plant Biotechnol J 20, 1648-1650.
DOI URL |
| [58] |
Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, Ulyanova AV, Yeldell SB, Griepenburg JC, Buckley PT, Kim J, Sul JY, Dmochowski IJ, Eberwine J (2014). Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods 11, 190-196.
DOI PMID |
| [59] |
Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L (2014). Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 11, 360-361.
DOI PMID |
| [60] |
Maïno N, Hauling T, Cappi G, Madaboosi N, Dupouy DG, Nilsson M (2019). A microfluidic platform towards automated multiplexed in situ sequencing. Sci Rep 9, 3542.
DOI PMID |
| [61] |
Maniatis S, Äijö T, Vickovic S, Braine C, Kang K, Mollbrink A, Fagegaltier D, Andrusivová Ž, Saarenpää S, Saiz-Castro G, Cuevas M, Watters A, Lundeberg J, Bonneau R, Phatnani H (2019). Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89-93.
DOI PMID |
| [62] |
Marks RA, Hotaling S, Frandsen PB, VanBuren R (2021). Representation and participation across 20 years of plant genome sequencing. Nat Plants 7, 1571-1578.
DOI PMID |
| [63] |
Marx V (2021). Method of the year: spatially resolved transcriptomics. Nat Methods 18, 9-14.
DOI PMID |
| [64] |
Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini II JL, Tran MN, Besich Z, Tippani M, Chew J, Yin YF, Kleinman JE, Hyde TM, Rao N, Hicks SC, Martinowich K, Jaffe AE (2021). Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci 24, 425-436.
DOI PMID |
| [65] |
Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li HJ, Iannacone M, Shulman Z, Amit I (2017). Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358, 1622-1626.
DOI PMID |
| [66] | Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, Nguyen K, Norgaard Z, Sorg K, Sprague I, Warren C, Warren S, Webster PJ, Zhou Z, Zollinger DR, Dunaway DL, Mills GB, Beechem JM (2020). Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 38, 586-599. |
| [67] |
Meylan M, Petitprez F, Becht E, Bougoüin A, Pupier G, Calvez A, Giglioli I, Verkarre V, Lacroix G, Verneau J, Sun CM, Laurent-Puig P, Vano YA, Elaïdi R, Méjean A, Sanchez-Salas R, Barret E, Cathelineau X, Oudard S, Reynaud CA, de Reyniès A, Sautès-Fridman C, Fridman WH (2022). Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity 55, 527-541.
DOI PMID |
| [68] |
Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, Hajdu CH, Simeone DM, Yanai I (2020). Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol 38, 333-342.
DOI PMID |
| [69] | Moreno-Villena JJ, Zhou HR, Gilman IS, Tausta SL, Cheung CYM, Edwards EJ (2022). Spatial resolution of an integrated C4+CAM photosynthetic metabolism. Sci Adv 8, eabn2349. |
| [70] |
Moses L, Pachter L (2022). Museum of spatial transcriptomics. Nat Methods 19, 534-546.
DOI PMID |
| [71] |
Nelms B, Walbot V (2019). Defining the developmental program leading to meiosis in maize. Science 364, 52-56.
DOI PMID |
| [72] | Okamura-Oho Y, Shimokawa K, Takemoto S, Hirakiyama A, Nakamura S, Tsujimura Y, Nishimura M, Kasukawa T, Masumoto KH, Nikaido I, Shigeyoshi Y, Ueda HR, Song G, Gee J, Himeno R, Yokota H (2012). Transcriptome tomography for brain analysis in the web-accessible anatomical space. PLoS One 7, e45373. |
| [73] | Orozco A (2020). A Spatial Analysis of Norwegian Spruce Cone Developmental Stages. Uppsala: Uppsala University. pp. 1-59. |
| [74] |
Ou ZH, Lin ST, Qiu JY, Ding WC, Ren PD, Chen DS, Wang JX, Tong YH, Wu D, Chen A, Deng Y, Cheng MN, Peng T, Lu HR, Yang HM, Wang J, Jin X, Ma D, Xu X, Wang YZ, Li JH, Wu P (2022). Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. Adv Sci 9, 2203040.
DOI URL |
| [75] |
Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V, Bejnood A, Dionne D, Ge WH, Xu KH, Chao SX, Zollinger DR, Lieb DJ, Reeves JW, Fuhrman CA, Hoang ML, Delorey T, Nguyen LT, Waldman J, Klapholz M, Wakiro I, Cohen O, Albers J, Smillie CS, Cuoco MS, Wu JY, Su MJ, Yeung J, Vijaykumar B, Magnuson AM, Asinovski N, Moll T, Goder-Reiser MN, Applebaum AS, Brais LK, DelloStritto LK, Denning SL, Phillips ST, Hill EK, Meehan JK, Frederick DT, Sharova T, Kanodia A, Todres EZ, Jané-Valbuena J, Biton M, Izar B, Lambden CD, Clancy TE, Bleday R, Melnitchouk N, Irani J, Kunitake H, Berger DL, Srivastava A, Hornick JL, Ogino S, Rotem A, Vigneau S, Johnson BE, Corcoran RB, Sharpe AH, Kuchroo VK, Ng K, Giannakis M, Nieman LT, Boland GM, Aguirre AJ, Anderson AC, Rozenblatt-Rosen O, Regev A, Hacohen N (2021). Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734-4752.
DOI PMID |
| [76] |
Peng GD, Cui GZ, Ke JC, Jing NH (2020). Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development. Annu Rev Genom Hum Genet 21, 163-181.
DOI URL |
| [77] |
Peng GD, Suo SB, Cui GZ, Yu F, Wang R, Chen J, Chen SR, Liu ZW, Chen GY, Qian Y, Tam PPL, Han JDJ, Jing NH (2019). Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572, 528-532.
DOI |
| [78] | Porritt RA, Zemmour D, Abe M, Lee Y, Narayanan M, Carvalho TT, Gomez AC, Martinon D, Santiskulvong C, Fishbein MC, Chen S, Crother TR, Shimada K, Arditi M, Rivas MN (2021). NLRP3 inflammasome mediates immune-stromal interactions in vasculitis. Circ Res 129, e183-e200. |
| [79] |
Potter SS (2018). Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 14, 479-492.
DOI PMID |
| [80] |
Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5, 877-879.
DOI PMID |
| [81] |
Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ (2019). Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463-1467.
DOI PMID |
| [82] |
Seyfferth C, Renema J, Wendrich JR, Eekhout T, Seurinck R, Vandamme N, Blob B, Saeys Y, Helariutta Y, Birnbaum KD, De Rybel B (2021). Advances and opportunities in single-cell transcriptomics for plant research. Annu Rev Plant Biol 72, 847-866.
DOI PMID |
| [83] |
Shah S, Lubeck E, Schwarzkopf M, He TF, Greenbaum A, Sohn CH, Lignell A, Choi HMT, Gradinaru V, Pierce NA, Cai L (2016). Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862-2867.
DOI PMID |
| [84] |
Shiroguchi K, Jia TZ, Sims PA, Xie XS (2012). Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci USA 109, 1347-1352.
DOI PMID |
| [85] |
Singer RH, Ward DC (1982). Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci USA 79, 7331-7335.
PMID |
| [86] |
Srivatsan SR, McFaline-Figueroa JL, Ramani V, Saunders L, Cao JY, Packer J, Pliner HA, Jackson DL, Daza RM, Christiansen L, Zhang F, Steemers F, Shendure J, Trapnell C (2020). Massively multiplex chemical transcriptomics at single-cell resolution. Science 367, 45-51.
DOI PMID |
| [87] |
Srivatsan SR, Regier MC, Barkan E, Franks JM, Packer JS, Grosjean P, Duran M, Saxton S, Ladd JJ, Spielmann M, Lois C, Lampe PD, Shendure J, Stevens KR, Trapnell C (2021). Embryo-scale, single-cell spatial transcriptomics. Science 373, 111-117.
DOI PMID |
| [88] |
Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78-82.
DOI PMID |
| [89] |
Stickels RR, Murray E, Kumar P, Li JL, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F (2021). Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seq V2. Nat Biotechnol 39, 313-319.
DOI PMID |
| [90] |
van den Brink SC, Alemany A, van Batenburg V, Moris N, Blotenburg M, Vivié J, Baillie-Johnson P, Nichols J, Sonnen KF, Martinez Arias A, van Oudenaarden A (2020). Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582, 405-409.
DOI |
| [91] |
Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, Gould J, Griffin GK, Borg Å, Ronaghi M, Frisén J, Lundeberg J, Regev A, Ståhl PL (2019). High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods 16, 987-990.
DOI PMID |
| [92] |
Wang MY, Hu QN, Lv TH, Wang YH, Lan Q, Xiang R, Tu ZC, Wei YR, Han K, Shi C, Guo JF, Liu C, Yang T, Du WS, An YR, Cheng MN, Xu JS, Lu HR, Li WS, Zhang SF, Chen A, Chen W, Li YX, Wang XS, Xu X, Hu YH, Liu LQ (2022). High-resolution 3D spatiotemporal transcriptomic maps of developing Drosophila embryos and larvae. Dev Cell 57, 1271-1283.
DOI URL |
| [93] | Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691. |
| [94] |
Wang Z, Portier BP, Gruver AM, Bui S, Wang HW, Su N, Vo HT, Ma XJ, Luo YL, Budd GT, Tubbs RR (2013). Automated quantitative RNA in situ hybridization for resolution of equivocal and heterogeneous ERBB2 (HER2) status in invasive breast carcinoma. J Mol Diagn 15, 210-219.
DOI URL |
| [95] |
Wei RM, He SY, Bai SS, Sei E, Hu M, Thompson A, Chen K, Krishnamurthy S, Navin NE (2022a). Spatial charting of single-cell transcriptomes in tissues. Nat Biotechnol 40, 1190-1199.
DOI |
| [96] | Wei XY, Fu SL, Li HB, Liu Y, Wang S, Feng WM, Yang YZ, Liu XW, Zeng YY, Cheng MN, Lai YW, Qiu XJ, Wu L, Zhang NN, Jiang YJ, Xu JS, Su XS, Peng C, Han L, Lou WPK, Liu CY, Yuan Y, Ma KL, Yang T, Pan XY, Gao S, Chen A, Esteban MA, Yang HM, Wang J, Fan GY, Liu LQ, Chen L, Xu X, Fei JF, Gu Y (2022b). Single-cell stereo-seq reveals induced progenitor cells involved in axolotl brain regeneration. Science 377, eabp9444. |
| [97] |
Weinstein JA, Regev A, Zhang F (2019). DNA microscopy: optics-free spatio-genetic imaging by a stand-alone chemical reaction. Cell 178, 229-241.
DOI PMID |
| [98] | Wendrich JR, Yang BJ, Vandamme N, Verstaen K, Smet W, van de Velde C, Minne M, Wybouw B, Mor E, Arents HE, Nolf J, Van Duyse J, Van Isterdael G, Maere S, Saeys Y, De Rybel B (2020). Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370, eaay4970. |
| [99] |
Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A (2022). An introduction to spatial transcriptomics for biomedical research. Genome Med 14, 68.
DOI PMID |
| [100] |
Wu DJ, Liu XZ, Zhang JQ, Li L, Wang XD (2021). Significance of single-cell and spatial transcriptomes in cell biology and toxicology. Cell Biol Toxicol 37, 1-5.
DOI PMID |
| [101] |
Xia KK, Sun HX, Li J, Li JM, Zhao Y, Chen LC, Qin C, Chen RY, Chen ZY, Liu GY, Yin RL, Mu BB, Wang XJ, Xu MY, Li XY, Yuan PS, Qiao YX, Hao SJ, Wang J, Xie Q, Xu JS, Liu SP, Li YX, Chen A, Liu LQ, Yin Y, Yang HM, Wang J, Gu Y, Xu X (2022). The single-cell stereo- seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves. Dev Cell 57, 1299-1310.
DOI URL |
| [102] |
Yao ZZ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, Ding SL, Fong O, Garren E, Glandon A, Gouwens NW, Gray J, Graybuck LT, Hawrylycz MJ, Hirschstein D, Kroll M, Lathia K, Lee C, Levi B, McMillen D, Mok S, Pham T, Ren QZ, Rimorin C, Shapovalova N, Sulc J, Sunkin SM, Tieu M, Torkelson A, Tung H, Ward K, Dee N, Smith KA, Tasic B, Zeng HK (2021). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222-3241.
DOI PMID |
| [103] | Zhang LL, Chen DS, Song DL, Liu XX, Zhang YN, Xu X, Wang XD (2022). Clinical and translational values of spatial transcriptomics. Signal Transduct Target Ther 7, 111. |
| [104] |
Zhang M, Eichhorn SW, Zingg B, Yao ZZ, Cotter K, Zeng HK, Dong HW, Zhuang XW (2021a). Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137-143.
DOI |
| [105] |
Zhang TQ, Chen Y, Liu Y, Lin WH, Wang JW (2021b). Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat Commun 12, 2053.
DOI |
| [1] | Niu Yanli, Bai Shenglong, Wang Qiyun, Liu Lingyun. Applications of Single-cell Technologies in Guard Cells [J]. Chinese Bulletin of Botany, 2017, 52(6): 788-796. |
| [2] | Zhigang Nie;Yan Wang;Shaoshan Li. Heavy Metal-induced DNA Damage in Arabidopsis thaliana Protoplasts Measured by Single-cell Gel Electrophoresis [J]. Chinese Bulletin of Botany, 2009, 44(01): 117-123. |
| [3] | Jing Wang;Lei Jiang;Yan Wang;Shaoshan Li. Sensitivity of Plant Leaves at Different Developmental Stages to UV-induced DNA Damage [J]. Chinese Bulletin of Botany, 2007, 24(02): 189-193. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||