

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (5): 584-593.DOI: 10.11983/CBB21104 cstr: 32102.14.CBB21104
• INVITED PROTOCOL • Previous Articles Next Articles
Jiayi Kuang, Hongqing Li, Wenjin Shen*( ), Caiji Gao*(
), Caiji Gao*( )
)
Received:2021-07-01
Accepted:2021-08-09
Online:2021-09-01
Published:2021-08-31
Contact:
Wenjin Shen,Caiji Gao
Jiayi Kuang, Hongqing Li, Wenjin Shen, Caiji Gao. Methods for TurboID-based Proximal Labeling in Plants[J]. Chinese Bulletin of Botany, 2021, 56(5): 584-593.
| Solution | Composition | Working concentration |
|---|---|---|
| Protein Extraction Buffer | Tris-HCl (pH7.5) | 50 mmol∙L-1 |
| NaCl | 150 mmol∙L-1 | |
| Sodium deoxycholate | 0.5% (w/v) | |
| SDS | 0.1% (w/v) | |
| EDTA | 1 mmol∙L-1 | |
| Triton X-100 | 1% (v/v) | |
| DTT | 1 mmol∙L-1 | |
| PMSF | 1 mmol∙L-1 | |
| Leupeptin | 10 μg∙mL-1 | |
| Protease Inhibitor Cocktail | 1× | |
| 4×SDS Sample Buffer | Tris-HCl (pH6.8) | 200 mmol∙L-1 |
| SDS | 8% (w/v) | |
| Glycerol | 40% (v/v) | |
| β-mercaptoethanol | 20% (v/v) | |
| Bromophenol blue | 0.1% (w/v) | |
| 1×PBS buffer (pH7.4) | NaH2PO4∙2H2O | 0.263 g∙L-1 |
| Na2HPO4∙12H2O | 1.856 g∙L-1 | |
| NaCl | 8 g∙L-1 | |
| KCl | 0.2 g∙L-1 | |
| 1×PBST | 1×PBS buffer containing 0.05% Tween-20 | |
| Biotin stock (stored at -20°C) | Dissolve biotin in DMSO to a final concentration of 50 mmol∙L-1 | |
Table 1 Related reagent formulations used in experiment
| Solution | Composition | Working concentration |
|---|---|---|
| Protein Extraction Buffer | Tris-HCl (pH7.5) | 50 mmol∙L-1 |
| NaCl | 150 mmol∙L-1 | |
| Sodium deoxycholate | 0.5% (w/v) | |
| SDS | 0.1% (w/v) | |
| EDTA | 1 mmol∙L-1 | |
| Triton X-100 | 1% (v/v) | |
| DTT | 1 mmol∙L-1 | |
| PMSF | 1 mmol∙L-1 | |
| Leupeptin | 10 μg∙mL-1 | |
| Protease Inhibitor Cocktail | 1× | |
| 4×SDS Sample Buffer | Tris-HCl (pH6.8) | 200 mmol∙L-1 |
| SDS | 8% (w/v) | |
| Glycerol | 40% (v/v) | |
| β-mercaptoethanol | 20% (v/v) | |
| Bromophenol blue | 0.1% (w/v) | |
| 1×PBS buffer (pH7.4) | NaH2PO4∙2H2O | 0.263 g∙L-1 |
| Na2HPO4∙12H2O | 1.856 g∙L-1 | |
| NaCl | 8 g∙L-1 | |
| KCl | 0.2 g∙L-1 | |
| 1×PBST | 1×PBS buffer containing 0.05% Tween-20 | |
| Biotin stock (stored at -20°C) | Dissolve biotin in DMSO to a final concentration of 50 mmol∙L-1 | |
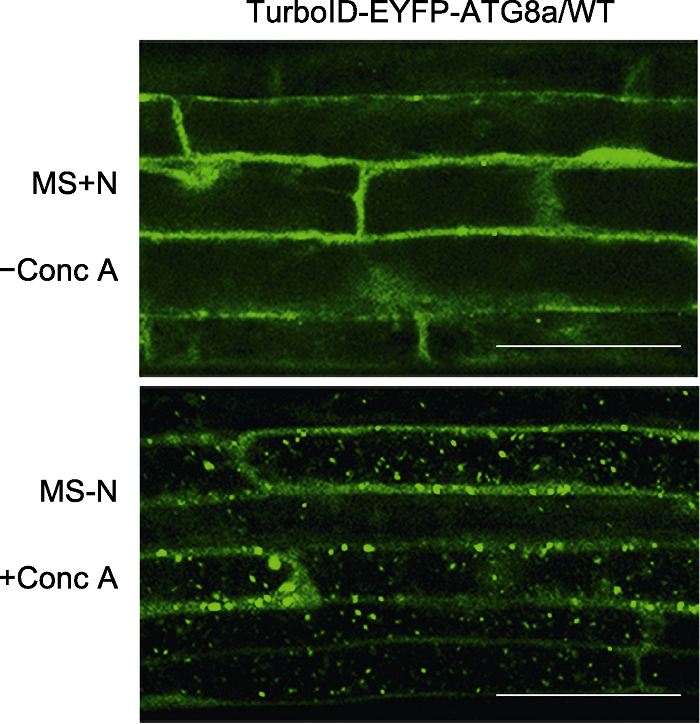
Figure 2 Subcellular localization of TurboID-EYFP-ATG8a in stable Arabidopsis lines 6-day-old transgenic Arabidopsis seedlings overexpressing TurboID-EYFP-ATG8a were transferred to nitrogen (N)- deficient medium with 1 µmol∙L-1 Conc A for 8 h followed by confocal microscopic analysis. Bars=50 μm
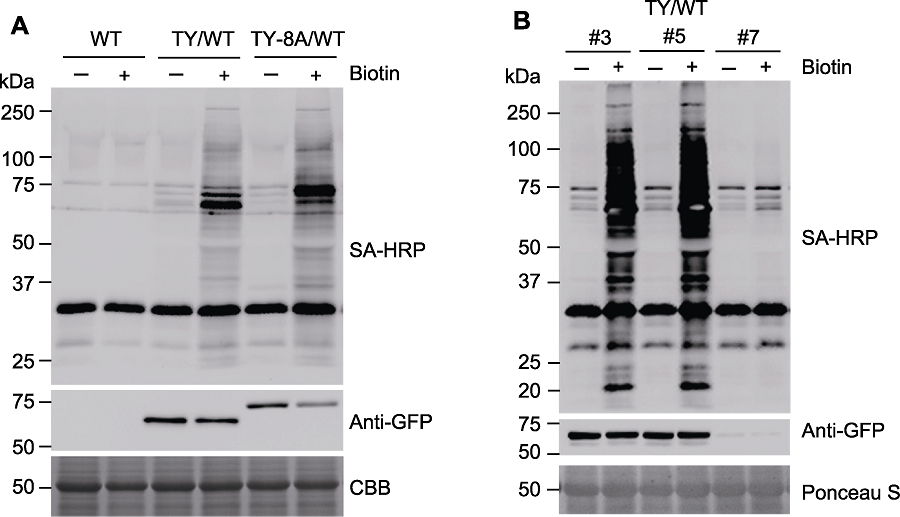
Figure 3 Immunoblot analysis of biotinylated protein (A) Wild-type (WT), transgenic Arabidopsis seedlings overexpressing TurboID-EYFP (TY/WT) or TurboID-EYFP-ATG8a (TY-8A/WT) were treated with 50 µmol∙L-1 biotin (+) for 3 h or 0 (-), activity and expression of TurboID-EYFP construct were analyzed by immunoblots with SA-HRP and anti-GFP antibodies; (B) Different lines of transgenic Arabidopsis seedlings overexpressing TurboID-EYFP (TY/WT) were treated with 50 µmol∙L-1 biotin (+) for 3 h or 0 (-), followed by immunoblots. SA-HRP: Streptavidin-HRP; CBB: Coomassie Brilliant Blue
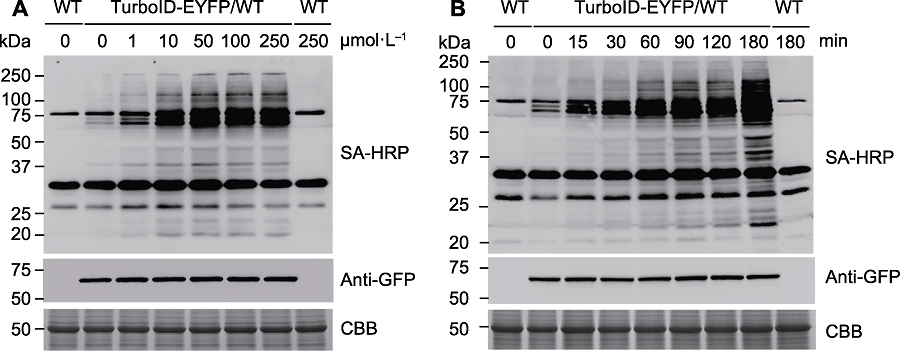
Figure 4 The optimization of biotin concentration and labeling time used in proximity labeling (A) Wild-type (WT) and transgenic Arabidopsis seedlings overexpressing TurboID-EYFP were treated with different concentrations of biotin solution for 1 h, followed by immunoblots with SA-HRP and anti-GFP antibodies to detect the proximity ligation activity and protein expression level of TurbolD-EYFP; (B) Wild-type (WT) and transgenic Arabidopsis seedings overexpressing TurboID-EYFP were treated with 50 µmol∙L-1 biotin for different times, followed by immunoblots with appropriate antibodies. SA-HRP: Streptavidin-HRP; CBB: Coomassie Brilliant Blue
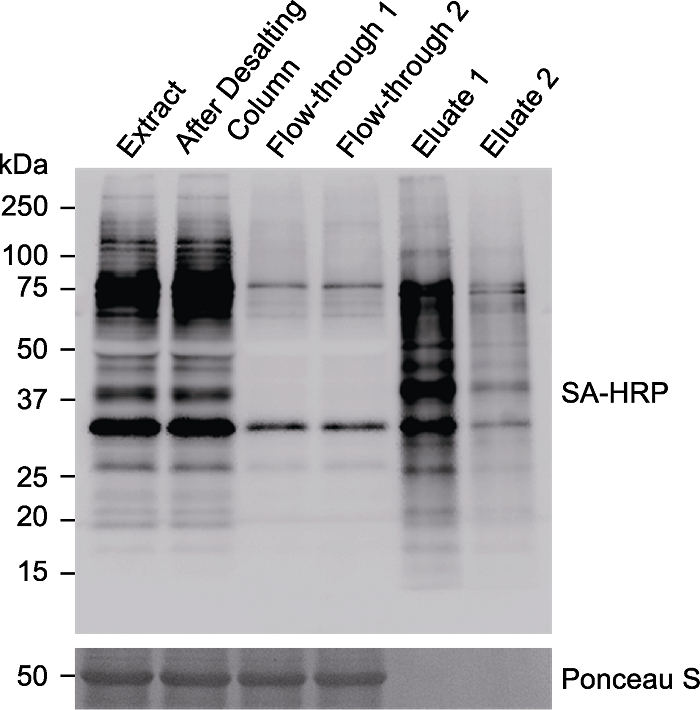
Figure 5 Immunoblotting analyses of the biotinylated proteins enriched by Streptavidin Magnetic Beads Arabidopsis seedlings overexpressing TurboID-EYFP-ATG8a were treated with 50 µmol∙L-1 biotin for 3 hours. The protein was extracted and desalted through Desalting Column. The desalted sample was split into two parts and incubated with 20 μL Streptavidin Magnetic Beads overnight, followed by protein elution with 20 μL or 200 μL 4× SDS Sample Buffer, respectively. Extract: Protein supernatant after centrifugation; After Desalting Column: Protein sample after desalting; Flow-through 1 or 2: Supernatant after overnight incubation with streptavidin (SA) beads; Eluate 1: Protein sample eluated with 20 μL buffer; Eluate 2: Protein sample eluated with 200 μL buffer; SA-HRP: Streptavidin-HRP
| [1] | 李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超 (2021). 植物细胞自噬基因的功能与转录调控机制. 植物学报 56, 201-217. |
| [2] | 刘洋, 张静, 王秋玲, 侯岁稳 (2018). 植物细胞自噬研究进展. 植物学报 53, 5-16. |
| [3] | 苏田, 韩笑, 刘华东 (2020). 邻近标记在蛋白质组学中的发展及应用. 中国生物化学与分子生物学报 36, 36-41. |
| [4] |
Alban C, Job D, Douce R (2000). Biotin metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol 51, 17-47.
DOI URL |
| [5] |
Arora D, Abel NB, Liu C, Van Damme P, Yperman K, Eeckhout D, Vu LD, Wang J, Tornkvist A, Impens F, Korbei B, Van Leene J, Goossens A, De Jaeger G, Ott T, Moschou PN, Van Damme D (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32, 3388-3407.
DOI URL |
| [6] |
Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880-887.
DOI PMID |
| [7] |
Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee HW, Lee IK, Carr SA, Ting AY (2020). Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci USA 117, 12143-12154.
DOI URL |
| [8] |
Kim TW, Park CH, Hsu CC, Zhu JY, Hsiao Y, Branon T, Xu SL, Ting AY, Wang ZY (2019). Application of TurboID mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. bioRxiv doi: https://doi.org/10.1101/636324.
DOI |
| [9] |
Lin QP, Zhou ZJ, Luo WB, Fang MC, Li MR, Li HQ (2017). Screening of proximal and interacting proteins in rice protoplasts by proximity-dependent biotinylation. Front Plant Sci 8, 749.
DOI URL |
| [10] |
Mair A, Xu SL, Branon TC, Ting AY, Bergmann DC (2019). Proximity labeling of protein complexes and cell- type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8, e47864.
DOI URL |
| [11] |
Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30, 1143-1148.
DOI URL |
| [12] |
Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801-810.
DOI URL |
| [13] |
Yang C, Shen WJ, Yang LM, Sun Y, Li XB, Lai MY, Wei J, Wang CJ, Xu YC, Li FQ, Liang S, Yang CW, Zhong SW, Luo M, Gao CJ (2020). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant 13, 515-531.
DOI URL |
| [14] |
Yang XX, Wen ZY, Zhang DL, Li Z, Li DW, Nagalakshmi U, Dinesh-Kumar SP, Zhang YL (2021). Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Commun 2, 100137.
DOI URL |
| [15] |
Zhang YL, Song GY, Lal NK, Nagalakshmi U, Li YY, Zheng WJ, Huang PJ, Branon TC, Ting AY, Walley JW, Dinesh-Kumar SP (2019). TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun 10, 3252.
DOI URL |
| [1] | Chongyi Xu. Pull-down and Co-immunoprecipitation Assays of Interacting Proteins in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 62-68. |
| [2] | Yan Zhao,Jianmin Zhou. Luciferase Complementation Assay for Detecting Protein Interactions [J]. Chinese Bulletin of Botany, 2020, 55(1): 69-75. |
| [3] | Lichao Chen,Ni Zhan,Yansha Li,Jian Feng,Jianru Zuo. Detection and Analysis of Protein S-nitrosylation in Plants [J]. Chinese Bulletin of Botany, 2019, 54(4): 497-502. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||