

拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响
收稿日期: 2023-06-05
录用日期: 2023-12-19
网络出版日期: 2023-12-29
基金资助
国家自然科学基金(32070216);山东省农业科学院农业科技创新工程(CXGC2023A15)
Effect of Amino Acid Point Mutations on the Structure and Function of Phytochrome B in Arabidopsis thaliana
Received date: 2023-06-05
Accepted date: 2023-12-19
Online published: 2023-12-29
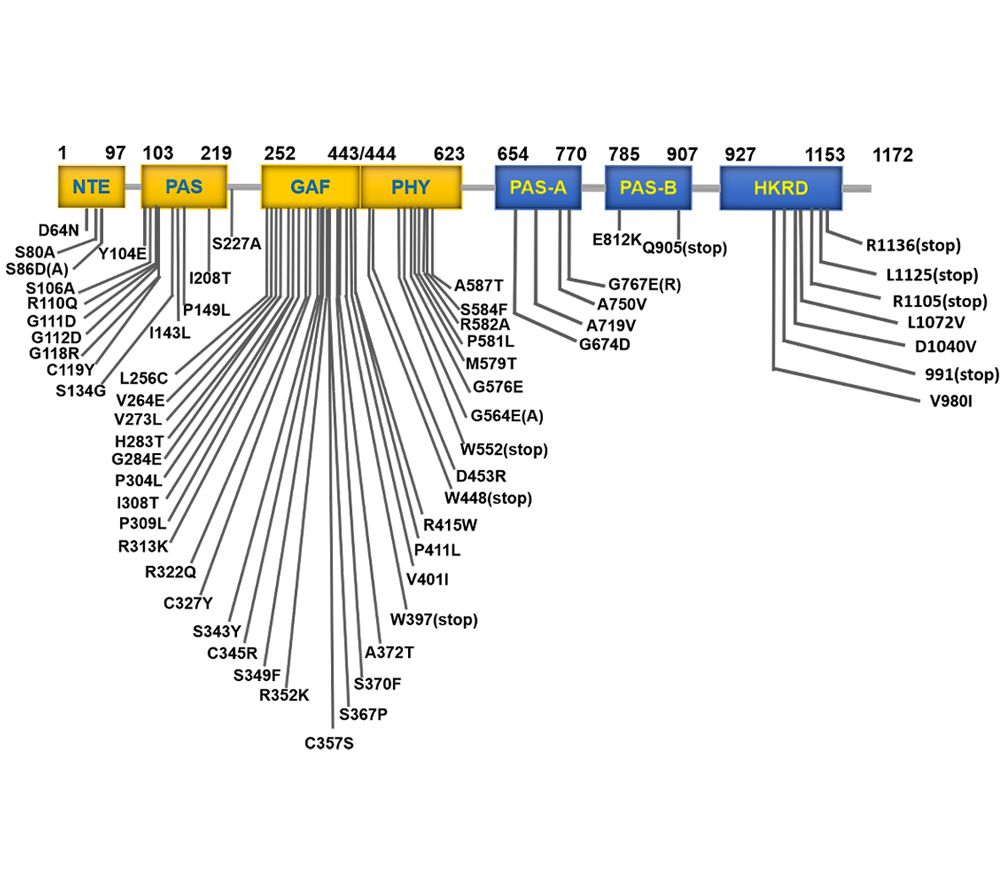
生物体为适应外界不断变化的光环境, 进化出不同的光受体, 其中光敏色素是一类经典的植物感受红光和远红光的受体蛋白, 其通过暗适应的Pr状态和光激活的Pfr状态之间的光转换来检测红光和远红光。植物光敏色素具有1个保守的N端感光区域和1个C端调节区域, 其中N端部分包括NTE、PAS、GAF和PHY亚结构域, C端部分包括2个PAS结构域和1个组氨酸激酶相关结构域(HKRD)。为深入了解光敏色素的结构及功能, 已获得许多光敏色素功能缺失或氨基酸位点突变体, 并对其进行功能研究, 发现N端结构域在光敏色素的光谱特性、光信号感知和光信号转导等方面均具有重要作用; 而C端结构域是光敏色素的二聚化与核定位所必需。该文综述了拟南芥(Arabidopsis thaliana)中光敏色素B (phyB)各亚结构域中氨基酸位点突变对其功能的影响, 以期深入理解phyB的结构及功能, 为未来通过基因编辑手段进行作物农艺性状的遗传改良奠定基础。
陈艳晓 , 李亚萍 , 周晋军 , 解丽霞 , 彭永彬 , 孙伟 , 和亚男 , 蒋聪慧 , 王增兰 , 郑崇珂 , 谢先芝 . 拟南芥光敏色素B氨基酸位点突变对其结构与功能的影响[J]. 植物学报, 2024 , 59(3) : 481 -494 . DOI: 10.11983/CBB23074
Organisms have evolved different photoreceptors to adapt to the ever-changing conditions of the external light environment. Phytochromes (phys) are one of the classic plant photoreceptors, mainly perceiving red and far-red light. Phytochromes detect red and far-red light through the light conversion between the dark-adapted Pr state and the light-activated Pfr state. All plant phytochromes have a conserved N-terminal photoreceptor region and a C-terminal regulatory region. The N-terminal includes NTE, PAS, GAF, and PHY subdomains, while C-terminal includes two PAS domains and a histidine kinase-related domain (HKRD). In order to understand how the structure of photochromes controls its function, many function-deficient photochrome derivatives and amino acid point mutants have been obtained and studied. The N-terminal domain plays important roles in the spectral properties, light signal perception and light signal transduction of phyB. The C-terminal domain is essential for dimerization and nuclear localization of photochrome. This paper mainly reviews point mutations of amino acid in various subdomains of phyB in Arabidopsis thaliana and their effects on the function of phyB, in order to have a better understanding of the structure and functional regulation of phyB. It lays a foundation for obtaining crops with desired agronomic characteristics through gene editing.

Key words: phyB; Arabidopsis thaliana; domain; amino acid point mutation; function
| [1] | 顾建伟, 刘婧, 薛彦久, 臧新, 谢先芝 (2011). 光敏色素在水稻生长发育中的作用. 中国水稻科学 25, 130-135. |
| [2] | 刘莎, 郑晓明, 冯雳, 乔卫华, 王君瑞, 公婷婷, 梁新霞, 齐兰, 苏龙, 丁膺宾, 许睿, 张丽芳, 程云连, 杨庆文 (2017). 水稻长日抑制基因PhyB的多样性及区域适应性初探. 植物遗传资源学报 18, 283-289. |
| [3] | 童哲, 赵玉锦, 王台, 李念华, 毛居代·亚力 (2000). 植物的光受体和光控发育研究. 植物学报 42, 111-115. |
| [4] | 张芳, 张晓枫, 王进征, 胡勇, 刘祥林 (2011). 植物光敏色素PHYA、PHYB研究进展. 生物学通报 46, 11-14. |
| [5] | 赵翔, 赵青平, 杨煦, 慕世超, 张骁 (2015). 向光素调节植物向光性及其与光敏色素/隐花色素的相互关系. 植物学报 50, 122-132. |
| [6] | ádám é, Hussong A, Bindics J, Wüst F, Viczián A, Essing M, Medzihradszky M, Kircher S, Sch?fer E, Nagy F (2011). Altered dark- and photo-conversion of phytochrome B mediate extreme light sensitivity and loss of photoreversibility of the phyB-401 mutant. PLoS One 6, e27250. |
| [7] | Anders K, Daminelli-Widany G, Mroginski MA, von Stetten D, Essen LO (2013). Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J Biol Chem 288, 35714-35725. |
| [8] | Auldridge ME, Forest KT (2011). Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol 46, 67-88. |
| [9] | Bae G, Choi G (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59, 281-311. |
| [10] | Bellini D, Papiz MZ (2012). Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436-1446. |
| [11] | Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD (2014). Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci USA 111, 10179-10184. |
| [12] | Burgie ES, Vierstra RD (2014). Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568-4583. |
| [13] | Chen D, Lyu M, Kou XX, Li J, Yang ZX, Gao LL, Li Y, Fan LM, Shi H, Zhong SW (2022). Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol Cell 82, 3015-3029. |
| [14] | Chen M, Chory J, Fankhauser C (2004). Light signal transduction in higher plants. Annu Rev Genet 38, 87-117. |
| [15] | Chen M, Schwab R, Chory J (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100, 14493-14498. |
| [16] | Chen M, Tao Y, Lim J, Shaw A, Chory J (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol 15, 637-642. |
| [17] | Cheng MC, Kathare PK, Paik I, Huq E (2021). Phytochrome signaling networks. Annu Rev Plant Biol 72, 217-244. |
| [18] | Cheng YL, Tu SL (2018). Alternative splicing and cross-talk with light signaling. Plant Cell Physiol 59, 1104-1110. |
| [19] | Cherry JR, Hondred D, Walker JM, Keller JM, Hershey HP, Vierstra RD (1993). Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell 5, 565-575. |
| [20] | Cherry JR, Hondred D, Walker JM, Vierstra RD (1992). Phytochrome requires the 6 kDa N-terminal domain for full biological activity. Proc Natl Acad Sci USA 89, 5039-5043. |
| [21] | Clack T, Mathews S, Sharrock RA (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25, 413-427. |
| [22] | Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA (2009). Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21, 786-799. |
| [23] | Dash L, McEwan RE, Montes C, Mejia L, Walley JW, Dilkes BP, Kelley DR (2021). Slim shady is a novel allele of PHYTOCHROME B present in the T-DNA line SALK_ 015201. Plant Direct 5, e00326. |
| [24] | Edgerton MD, Jones AM (1992). Localization of protein- protein interactions between subunits of phytochrome. Plant Cell 4, 161-171. |
| [25] | Elich TD, Chory J (1997). Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell 9, 2271-2280. |
| [26] | Essen LO, Mailliet J, Hughes J (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA 105, 14709-14714. |
| [27] | Franklin KA, Quail PH (2010). Phytochrome functions in Arabidopsis development. J Exp Bot 61, 11-24. |
| [28] | He YN, Li YP, Cui LX, Xie LX, Zheng CK, Zhou GH, Zhou JJ, Xie XZ (2016). Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice. Front Plant Sci 7, 1963. |
| [29] | Hernando CE, Murcia MG, Pereyra ME, Sellaro R, Casal JJ (2021). Phytochrome B links the environment to transcription. J Exp Bot 72, 4068-4084. |
| [30] | Hu W, Su YS, Lagarias JC (2009). A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2, 166-182. |
| [31] | Jung JH, Li Z, Chen H, Yang S, Li DD, Priatama RA, Kumar V, Xuan YH (2023). Mutation of phytochrome B promotes resistance to sheath blight and saline-alkaline stress via increasing ammonium uptake in rice. Plant J 113, 277-290. |
| [32] | Kikis EA, Oka Y, Hudson ME, Nagatani A, Quail PH (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet 5, e1000352. |
| [33] | Kim C, Kwon Y, Jeong J, Kang MJ, Lee GS, Moon JH, Lee HJ, Park YI, Choi G (2023). Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat Commun 14, 1708. |
| [34] | Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, ádám é, Sch?fer E, Nagy F (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14, 1541-1555. |
| [35] | Klose C, Venezia F, Hussong A, Kircher S, Sch?fer E, Fleck C (2015). Systematic analysis of how phytochrome B dimerization determines its specificity. Nat Plants 1, 15090. |
| [36] | Krall L, Reed JW (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA 97, 8169-8174. |
| [37] | Kretsch T, Poppe C, Sch?fer E (2000). A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J 22, 177-186. |
| [38] | Kwon CT, Song G, Kim SH, Han J, Yoo SC, An G, Kang K, Paek NC (2018). Functional deficiency of phytochrome B improves salt tolerance in rice. Environ Exp Bot 148, 100-108. |
| [39] | Legris M, Ince Y?, Fankhauser C (2019). Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun 10, 5219. |
| [40] | Li H, Burgie ES, Gannam ZTK, Li HL, Vierstra RD (2022). Plant phytochrome B is an asymmetric dimer with unique signaling potential. Nature 604, 127-133. |
| [41] | Liu J, Zhang F, Zhou JJ, Chen F, Wang BS, Xie XZ (2012). Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol Biol 78, 289-300. |
| [42] | Liu X, Jiang W, Li YL, Nie HZ, Cui LN, Li RX, Tan L, Peng L, Li C, Luo JY, Li M, Wang HX, Yang J, Zhou B, Wang PC, Liu HT, Zhu JK, Zhao CZ (2023). FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat Plants 9, 645-660. |
| [43] | Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, Weigel D, Chory J (2001). Natural variation in light sensitivity of Arabidopsis. Nat Genet 29, 441-446. |
| [44] | Mathews S (2006). Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol 15, 3483-3503. |
| [45] | Matsushita T, Mochizuki N, Nagatani A (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571-574. |
| [46] | Medzihradszky M, Bindics J, ádám é, Viczián A, Klement é, Lorrain S, Gyula P, Mérai Z, Fankhauser C, Medzihradszky KF, Kunkel T, Sch?fer E, Nagy F (2013). Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25, 535-544. |
| [47] | Nagano S (2016). From photon to signal in phytochromes: similarities and differences between prokaryotic and plant phytochromes. J Plant Res 129, 123-135. |
| [48] | Nito K, Wong CCL, Yates III JR, Chory J (2013). Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep 3, 1970-1979. |
| [49] | Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A (2008). Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet 4, e1000158. |
| [50] | Oka Y, Matsushita T, Mochizuki N, Suzuki T, Tokutomi S, Nagatani A (2004). Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16, 2104-2116. |
| [51] | Paik I, Huq E (2019). Plant photoreceptors: multi-functional sensory proteins and their signaling networks. Semin Cell Dev Biol 92, 114-121. |
| [52] | Qiu YJ, Pasoreck EK, Reddy AK, Nagatani A, Ma WX, Chory J, Chen M (2017). Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat Commun 8, 1905. |
| [53] | Quail PH (2002). Phytochrome photosensory signaling networks. Nat Rev Mol Cell Biol 3, 85-93. |
| [54] | Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147-157. |
| [55] | Rockwell NC, Lagarias JC (2006). The structure of phytochrome: a picture is worth a thousand spectra. Plant Cell 18, 4-14. |
| [56] | Rockwell NC, Su YS, Lagarias JC (2006). Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57, 837-858. |
| [57] | Ruiz-Diaz MJ, Matsusaka D, Cascales J, Sánchez DH, Sánchez-Lamas M, Cerdán PD, Botto JF (2022). Functional analysis of PHYB polymorphisms in Arabidopsis thaliana collected in Patagonia. Front Plant Sci 13, 952214. |
| [58] | Sharrock RA, Quail PH (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3, 1745-1757. |
| [59] | Su YS, Lagarias JC (2007). Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19, 2124-2139. |
| [60] | Sun W, Xu XH, Lu XB, Xie LX, Bai B, Zheng CK, Sun HW, He YN, Xie XZ (2017). The rice phytochrome genes, PHYA and PHYB, have synergistic effects on anther development and pollen viability. Sci Rep 7, 6439. |
| [61] | Velázquez Escobar F, Buhrke D, Fernandez Lopez M, Shenkutie SM, von Horsten S, Essen LO, Hughes J, Hildebrandt P (2017). Structural communication between the chromophore-binding pocket and the N-terminal extension in plant phytochrome phyB. FEBS Lett 591, 1258-1265. |
| [62] | Viczián A, ádám é, Staudt AM, Lambert D, Klement E, Romero Montepaone S, Hiltbrunner A, Casal J, Sch?fer E, Nagy F, Klose C (2020). Differential phosphorylation of the N-terminal extension regulates phytochrome B signaling. New Phytol 225, 1635-1650. |
| [63] | Viczián A, Nagy F (2024). Phytochrome B phosphorylation expanded: site-specific kinases are identified. New Phytol 241, 65-72. |
| [64] | Vierstra RD (1993). Illuminating phytochrome functions (There is light at the end of the tunnel). Plant Physiol 103, 679-684. |
| [65] | Wagner D, Koloszvari M, Quail PH (1996). Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8, 859-871. |
| [66] | Wagner D, Quail PH (1995). Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc Natl Acad Sci USA 92, 8596-8600. |
| [67] | Wagner JR, Brunzelle JS, Forest KT, Vierstra RD (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325-331. |
| [68] | Wagner JR, Zhang JR, Brunzelle JS, Vierstra RD, Forest KT (2007). High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem 282, 12298-12309. |
| [69] | Wies G, Mantese AI, Casal JJ, Maddonni Gá (2019). Phytochrome B enhances plant growth, biomass and grain yield in field-grown maize. Ann Bot 123, 1079-1088. |
| [70] | Xie XZ, Shinomura T, Inagaki N, Kiyota S, Takano M (2007). Phytochrome-mediated inhibition of coleoptile growth in rice: age-dependency and action spectra. Photochem Photobiol 83, 131-138. |
| [71] | Xie XZ, Xue YJ, Zhou JJ, Zhang B, Chang H, Takano M (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol Plant 4, 688-696. |
| [72] | Xu Y, Parks BM, Short TW, Quail PH (1995). Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7, 1433-1443. |
| [73] | Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145, 437-445. |
| [74] | Yang XJ, Kuk J, Moffat K (2008). Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci USA 105, 14715-14720. |
| [75] | Yang XJ, Kuk J, Moffat K (2009). Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc Natl Acad Sci USA 106, 15639-15644. |
| [76] | Zhang JR, Stankey RJ, Vierstra RD (2013). Structure- guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol 161, 1445-1457. |
| [77] | Zhao Y, Shi H, Pan Y, Lyu M, Yang ZX, Kou XX, Deng XW, Zhong SW (2023). Sensory circuitry controls cytosolic calcium-mediated phytochrome B phototransduction. Cell 186, 1230-1243. |
/
| 〈 |
|
〉 |