

水稻穗粒数遗传机制与育种利用
收稿日期: 2023-02-04
录用日期: 2023-04-18
网络出版日期: 2023-04-28
基金资助
国家自然科学基金(32071993)
Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice
Received date: 2023-02-04
Accepted date: 2023-04-18
Online published: 2023-04-28
严语萍, 俞晓琦, 任德勇, 钱前 . 水稻穗粒数遗传机制与育种利用[J]. 植物学报, 2023 , 58(3) : 359 -372 . DOI: 10.11983/CBB23012
As one of the key factors affecting rice yield, the grain number per panicle has always attracted the attention of breeders. The formation of grain number per panicle is a complex biological process, which is regulated by many genes. According to their impact on phenotype, we roughly divide these genes into three categories: related to the number of branches, related to panicle type and related to spikelet determination. In this paper, we summarized the genetic regulation mechanisms of the grain number per panicle-related genes, and put forth the strategies for their use in high yield breeding of rice.
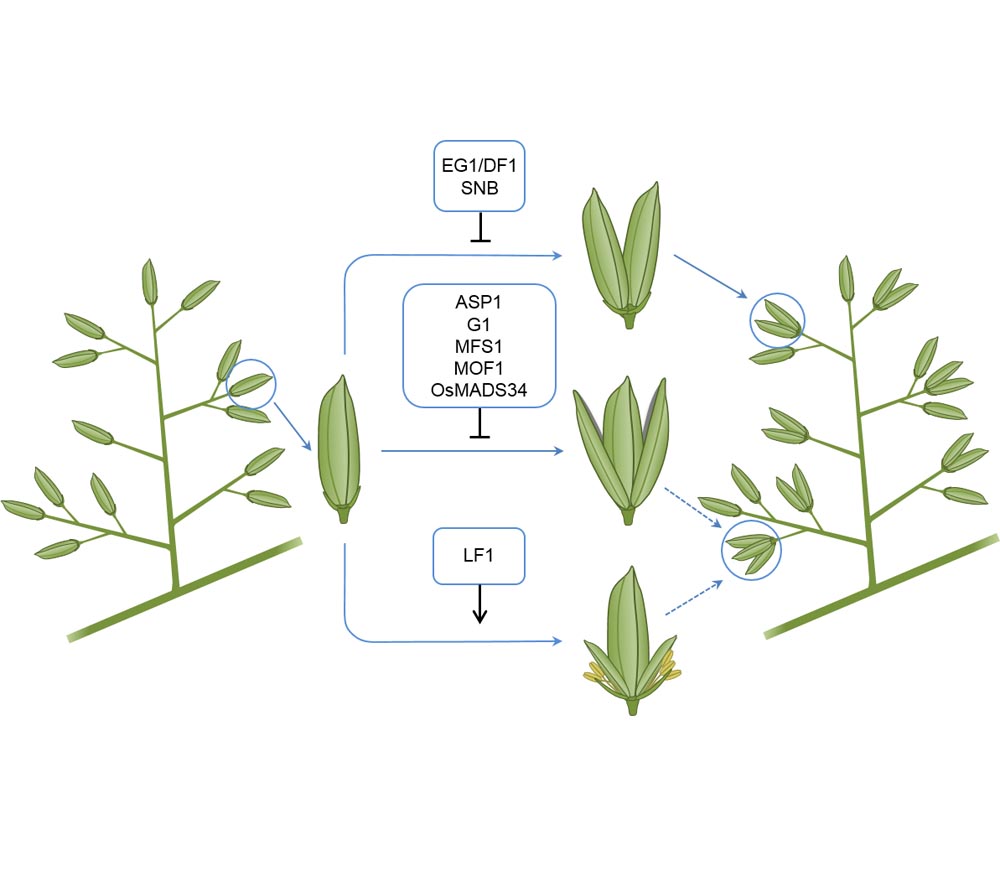
Key words: rice; yield; grain number per panicle; genetic mechanism; breeding
| [1] | 任德勇, 何光华, 凌英华, 桑贤春, 杨正林, 赵芳明 (2010). 基于单片段代换系的水稻穗长QTL加性及其上位性效应. 植物学报 45, 662-669. |
| [2] | 尚江源, 淳雁, 李学勇 (2021). 水稻穗长基因PAL3的克隆及自然变异分析. 植物学报 56, 520-532. |
| [3] | 杨凯如, 贾绮玮, 金佳怡, 叶涵斐, 王盛, 陈芊羽, 管易安, 潘晨阳, 辛德东, 方媛, 王跃星, 饶玉春 (2022). 水稻黄绿叶调控基因YGL18的克隆与功能解析. 植物学报 57, 276-287. |
| [4] | 周亭亭, 饶玉春, 任德勇 (2018). 水稻卷叶细胞学与分子机制研究进展. 植物学报 53, 848-855. |
| [5] | 朱克明, 陶慧敏, 闵超, 杨艳华 (2015). 水稻穗发育调控分子遗传研究进展. 分子植物育种 13, 2109-2117. |
| [6] | Ali A, Xu PZ, Riaz A, Wu XJ (2019). Current advances in molecular mechanisms and physiological basis of panicle degeneration in rice. Int J Mol Sci 20, 1613. |
| [7] | Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741-745. |
| [8] | Bai XF, Huang Y, Hu Y, Liu HY, Zhang B, Smaczniak C, Hu G, Han ZM, Xing YZ (2017). Duplication of an upstream silencer of FZP increases grain yield in rice. Nat Plants 3, 885-893. |
| [9] | Chen H, Tang YY, Liu JF, Tan LB, Jiang JH, Wang MM, Zhu ZF, Sun XY, Sun CQ (2017). Emergence of a novel chimeric gene underlying grain number in rice. Genetics 205, 993-1002. |
| [10] | Chu HW, Qian Q, Liang WQ, Yin CS, Tan HX, Yao X, Yuan Z, Yang J, Huang H, Luo D, Ma H, Zhang DB (2006). The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142, 1039-1052. |
| [11] | Dai ZY, Wang J, Yang XF, Lu H, Miao XX, Shi ZY (2018). Modulation of plant architecture by the miR156f-OsSPL7- OsGH3.8 pathway in rice. J Exp Bot 69, 5117-5130. |
| [12] | Deveshwar P, Prusty A, Sharma S, Tyagi AK (2020). Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front Genet 11, 586462. |
| [13] | Duan EC, Wang YH, Li XH, Lin QB, Zhang T, Wang YP, Zhou CL, Zhang H, Jiang L, Wang JL, Lei CL, Zhang X, Guo XP, Wang HY, Wan JM (2019). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026-1042. |
| [14] | Dun EA, De Saint Germain A, Rameau C, Beveridge CA, (2012). Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158, 487-498. |
| [15] | Feng XM, Wang C, Nan JZ, Zhang XH, Wang RS, Jiang GQ, Yuan QB, Lin SY (2017). Updating the elite rice variety Kongyu 131 by improving the Gn1a locus. Rice 10, 35. |
| [16] | Gao F, Wang K, Liu Y, Chen YP, Chen P, Shi ZY, Luo J, Jiang DQ, Fan FF, Zhu YG, Li SQ (2016). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat Plants 2, 15196. |
| [17] | Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871-888. |
| [18] | Guo T, Lu ZQ, Shan JX, Ye WW, Dong NQ, Lin HX (2020). ERECTA1 acts upstream of the OsMKKK10-OsMKK4- OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell 32, 2763-2779. |
| [19] | He YB, Yan L, Ge CN, Yao XF, Han X, Wang RC, Xiong LZ, Jiang LW, Liu CM, Zhao YD (2019). PINOID is required for formation of the stigma and style in rice. Plant Physiol 180, 926-936. |
| [20] | Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal- Bertioli SCM, Tester M, Gao CX, Godwin ID, Hayes BJ, Wulff BBH, (2019). Breeding crops to feed 10 billion. Nat Biotechnol 37, 744-754. |
| [21] | Huang XZ, Qian Q, Liu ZB, Sun HY, He SY, Luo D, Xia GM, Chu CC, Li JY, Fu XD (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41, 494-497. |
| [22] | Huang Y, Bai XF, Luo MF, Xing YZ (2019). Short Panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice. J Integr Plant Biol 61, 987-999. |
| [23] | Huang YY, Zhao SS, Fu YC, Sun HD, Ma X, Tan LB, Liu FX, Sun XY, Sun HY, Gu P, Xie DX, Sun CQ, Zhu ZF (2018). Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J 96, 716-733. |
| [24] | Huo X, Wu S, Zhu ZF, Liu FX, Fu YC, Cai HW, Sun XY, Gu P, Xie DX, Tan LB, Sun CQ (2017). NOG1 increases grain production in rice. Nat Commun 8, 1497. |
| [25] | Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51, 1030-1040. |
| [26] | Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh JI, Nagato Y (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69, 168-180. |
| [27] | Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15, 1560-1565. |
| [28] | Jiang DG, Chen WT, Dong JF, Li J, Yang F, Wu ZC, Zhou H, Wang WS, Zhuang CX (2018). Overexpression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J Exp Bot 69, 1533-1543. |
| [29] | Jiang GH, Xiang YH, Zhao JY, Yin DD, Zhao XF, Zhu LH, Zhai WX (2014). Regulation of inflorescence branch development in rice through a novel pathway involving the pentatricopeptide repeat protein sped1-D. Genetics 197, 1395-1407. |
| [30] | Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD, Qian Q, Li JY (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42, 541-544. |
| [31] | Kim SR, Ramos JM, Hizon RJM, Ashikari M, Virk PS, Torres EA, Nissila E, Jena KK (2018). Introgression of a functional epigenetic OsSPL14WFP allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci Rep 8, 3833. |
| [32] | Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51, 47-57. |
| [33] | Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003a). LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100, 11765-11770. |
| [34] | Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003b). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841-3850. |
| [35] | Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652-655. |
| [36] | Kwon Y, Yu SI, Park JH, Li Y, Han JH, Alavilli H, Cho JI, Kim TH, Jeon JS, Lee BH (2012). OsREL2, a rice TOPLESS homolog functions in axillary meristem development in rice inflorescence. Plant Biotechnol Rep 6, 213-224. |
| [37] | Kyozuka J (2007). Control of shoot and root meristem function by cytokinin. Curr Opin Plant Biol 10, 442-446. |
| [38] | Lee DY, An G (2012). Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKE- LET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69, 445-461. |
| [39] | Lee DY, Lee J, Moon S, Park SY, An G (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49, 64-78. |
| [40] | Li GL, Xu BX, Zhang YP, Xu YW, Khan NU, Xie JY, Sun XM, Guo HF, Wu ZY, Wang XQ, Zhang HL, Li JJ, Xu JL, Wang WS, Zhang ZY, Li ZC (2022). RGN1 controls grain number and shapes panicle architecture in rice. Plant Biotechnol J 20, 158-167. |
| [41] | Li HG, Xue DW, Gao ZY, Yan MX, Xu WY, Xing Z, Huang DN, Qian Q, Xue YB (2009a). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J 57, 593-605. |
| [42] | Li M, Tang D, Wang KJ, Wu XR, Lu LL, Yu HX, Gu MH, Yan CJ, Cheng ZK (2011). Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J 9, 1002-1013. |
| [43] | Li SB, Qian Q, Fu ZM, Zeng DL, Meng XB, Kyozuka J, Maekawa M, Zhu XD, Zhang J, Li JY, Wang YH (2009b). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58, 592-605. |
| [44] | Li SY, Zhao BR, Yuan DY, Duan MJ, Qian Q, Tang L, Wang B, Liu XQ, Zhang J, Wang J, Sun JQ, Liu Z, Feng YQ, Yuan LP, Li CY (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/ OsCKX2 expression. Proc Natl Acad Sci USA 110, 3167-3172. |
| [45] | Liao ZG, Yu H, Duan JB, Yuan K, Yu CJ, Meng XB, Kou LQ, Chen MJ, Jing YH, Liu GF, Smith SM, Li JY (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat Commun 10, 2738. |
| [46] | Liu HH, Guo SY, Xu YY, Li CH, Zhang ZY, Zhang DJ, Xu SJ, Zhang C, Chong K (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Phy-siol 165, 160-174. |
| [47] | Lu ZF, Yu H, Xiong GS, Wang J, Jiao YQ, Liu GF, Jing YH, Meng XB, Hu XM, Qian Q, Fu XD, Wang YH, Li JY (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743-3759. |
| [48] | Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42, 545-549. |
| [49] | Mjomba FM, Zheng Y, Liu HQ, Tang WQ, Hong ZL, Wang F, Wu WR (2016). Homeobox is pivotal for OsWUS controlling tiller development and female fertility in rice. G3 6, 2013-2021. |
| [50] | Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, Ebitani T, Ozaki H, Angeles ER, Hirasawa T, Matsuoka M (2010). New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1, 132. |
| [51] | Piao RH, Jiang WZ, Ham TH, Choi MS, Qiao YL, Chu SH, Park JH, Woo MO, Jin ZX, An G, Lee J, Koh HJ (2009). Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor Appl Genet 119, 1497-1506. |
| [52] | Qiao YL, Piao RH, Shi JX, Lee SI, Jiang WZ, Kim BK, Lee J, Han LZ, Ma WB, Koh HJ (2011). Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor Appl Genet 122, 1439-1449. |
| [53] | Ren DY, Li YF, Zhao FM, Sang XC, Shi JQ, Wang N, Guo S, Ling YH, Zhang CW, Yang ZL, He GH (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol 162, 872-884. |
| [54] | Ren DY, Rao YC, Yu HP, Xu QK, Cui YJ, Xia SS, Yu XQ, Liu H, Hu HT, Xue DW, Zeng DL, Hu J, Zhang GH, Gao ZY, Zhu L, Zhang Q, Shen L, Guo LB, Qian Q (2020). MORE FLORET1 encodes a MYB transcription factor that regulates spikelet development in rice. Plant Physiol 184, 251-265. |
| [55] | Ren DY, Xu QK, Qiu ZN, Cui YJ, Zhou TT, Zeng DL, Guo LB, Qian Q (2019). FON4 prevents the multi-floret spikelet in rice. Plant Biotechnol J 17, 1007-1009. |
| [56] | Ren DY, Yu HP, Rao YC, Xu QK, Zhou TT, Hu J, Zhang Y, Zhang GH, Zhu L, Gao ZY, Chen G, Guo LB, Zeng DL, Qian Q (2018). 'Two-floret spikelet' as a novel resource has the potential to increase rice yield. Plant Biotechnol J 16, 351-353. |
| [57] | Shao GN, Lu ZF, Xiong JS, Wang B, Jing YH, Meng XB, Liu GF, Ma HY, Liang Y, Chen F, Wang YH, Li JY, Yu H (2019). Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol Plant 12, 1090-1102. |
| [58] | Si LZ, Chen JY, Huang XH, Gong H, Luo JH, Hou QQ, Zhou TY, Lu TT, Zhu JJ, Shangguan YY, Chen EW, Gong CX, Zhao Q, Jing YF, Zhao Y, Li Y, Cui LL, Fan DL, Lu YQ, Weng QJ, Wang YC, Zhan QL, Liu KY, Wei XH, An K, An G, Han B (2016). OsSPL13 controls grain size in cultivated rice. Nat Genet 48, 447-456. |
| [59] | Song S, Wang GF, Hu Y, Liu HY, Bai XF, Qin R, Xing YZ (2018). OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J Exp Bot 69, 4283-4293. |
| [60] | Suzuki C, Tanaka W, Hirano HY (2019). Transcriptional corepressor ASP1 and CLV-like signaling regulate meristem maintenance in rice. Plant Physiol 180, 1520-1534. |
| [61] | Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131, 5649-5657. |
| [62] | Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol 47, 1591-1602. |
| [63] | Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang XH, Yoshida H, Kyozuka J, Chen F, Sato Y (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276-3287. |
| [64] | Tang L, Xu ZJ, Chen WF (2017). Advances and prospects of super rice breeding in China. J Integr Agric 16, 984-991. |
| [65] | Tsuda K, Ito Y, Sato Y, Kurata N (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368-4381. |
| [66] | Wang B, Smith SM, Li JY (2018). Genetic regulation of shoot architecture. Annu Rev Plant Biol 69, 437-468. |
| [67] | Wang J, Yu H, Xiong GS, Lu ZF, Jiao YQ, Meng XB, Liu GF, Chen XW, Wang YH, Li JY (2017a). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29, 697-707. |
| [68] | Wang L, Sun SY, Jin JY, Fu DB, Yang XF, Weng XY, Xu CG, Li XH, Xiao JH, Zhang QF (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci USA 112, 15504-15509. |
| [69] | Wang SS, Wu K, Qian Q, Liu Q, Li Q, Pan YJ, Ye YF, Liu XY, Wang J, Zhang JQ, Li S, Wu YJ, Fu XD (2017b). Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res 27, 1142-1156. |
| [70] | Wu HM, Xie DJ, Tang ZS, Shi DQ, Yang WC (2020). PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice. Plant Biotechnol J 18, 1778-1795. |
| [71] | Wu Y, Wang Y, Mi XF, Shan JX, Li XM, Xu JL, Lin HX (2016). The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet 12, e1006386. |
| [72] | Xing YZ, Zhang QF (2010). Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61, 421-442. |
| [73] | Xu C, Wang YH, Yu YC, Duan JB, Liao ZG, Xiong GS, Meng XB, Liu GF, Qian Q, Li JY (2012). Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat Commun 3, 750. |
| [74] | Xu QK, Yu XQ, Cui YJ, Xia SS, Zeng DL, Qian Q, Ren DY (2021). LRG1 maintains sterile lemma identity by regulating OsMADS6 expression in rice. Sci China Life Sci 64, 1190-1192. |
| [75] | Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano HY (2012). ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-re- pressor, is involved in the regulation of meristem fate in rice. Plant J 70, 327-339. |
| [76] | Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen RH, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, Nagamura Y, Ushijima T, Kumamaru T, Iida S, Maekawa M, Kyozuka J (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110, 767-772. |
| [77] | Yoshida A, Suzaki T, Tanaka W, Hirano HY (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA 106, 20103-20108. |
| [78] | Yoshida H, Nagato Y (2011). Flower development in rice. J Exp Bot 62, 4719-4730. |
| [79] | Yuan H, Qin P, Hu L, Zhan SJ, Wang SF, Gao P, Li J, Jin MY, Xu ZY, Gao Q, Du AP, Tu B, Chen WL, Ma BT, Wang YP, Li SG (2019). OsSPL18 controls grain weight and grain number in rice. J Genet Genomics 46, 41-51. |
| [80] | Yuan H, Xu ZY, Tan XQ, Gao P, Jin MY, Song WC, Wang SG, Kang YH, Liu PX, Tu B, Wang YP, Qin P, Li SG, Ma BT, Chen WL (2021). A natural allele of TAW1 contributes to high grain number and grain yield in rice. Crop J 9, 1060-1069. |
| [81] | Zhang GH, Li SY, Wang L, Ye WJ, Zeng DL, Rao YC, Peng YL, Hu J, Yang YL, Xu J, Ren DY, Gao ZY, Zhu L, Dong GJ, Hu XM, Yan MX, Guo LB, Li CY, Qian Q (2014). LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol Plant 7, 1350-1364. |
| [82] | Zhang T, Li YF, Ma L, Sang XC, Ling YH, Wang YT, Yu P, Zhuang H, Huang JY, Wang N, Zhao FM, Zhang CW, Yang ZL, Fang LK, He GH (2017). LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc Natl Acad Sci USA 114, 9984-9989. |
| [83] | Zhang Y, Yu HP, Liu J, Wang W, Sun J, Gao Q, Zhang YH, Ma DR, Wang JY, Xu ZJ, Chen WF (2016). Loss of function of OsMADS34 leads to large sterile lemma and low grain yield in rice (Oryza sativa L.). Mol Breeding 36, 147. |
| [84] | Zhang ZY, Li JJ, Tang ZS, Sun XM, Zhang HL, Yu JP, Yao GX, Li GL, Guo HF, Li JL, Wu HM, Huang HG, Xu YW, Yin ZG, Qi YH, Huang RF, Yang WC, Li ZC (2018). Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3- OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J Exp Bot 69, 4723-4737. |
| [85] | Zhang ZY, Sun XM, Ma XQ, Xu BX, Zhao Y, Ma ZQ, Li GL, Khan NU, Pan YH, Liang YT, Zhang HL, Li JJ, Li ZC (2021). GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J 9, 57-67. |
| [86] | Zhu KM, Tang D, Yan CJ, Chi ZC, Yu HX, Chen JM, Liang JS, Gu MH, Cheng ZK (2010). ERECT PANICLE2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics 184, 343-350. |
| [87] | Zhu WW, Yang L, Wu D, Meng QC, Deng X, Huang GQ, Zhang J, Chen XF, Ferrándiz C, Liang WQ, Dreni L, Zhang DB (2022). Rice SEPALLATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4. New Phytol 233, 1682-1700. |
/
| 〈 |
|
〉 |