

甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用
收稿日期: 2022-04-17
录用日期: 2022-07-03
网络出版日期: 2022-07-04
基金资助
国家自然科学基金(31971836);湖南省教育厅优秀青年科学基金(19B279)
Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance
Received date: 2022-04-17
Accepted date: 2022-07-03
Online published: 2022-07-04
吴楠 , 覃磊 , 崔看 , 李海鸥 , 刘忠松 , 夏石头 . 甘蓝型油菜EXA1的克隆及其对植物抗病的调控作用[J]. 植物学报, 2023 , 58(3) : 385 -393 . DOI: 10.11983/CBB22077
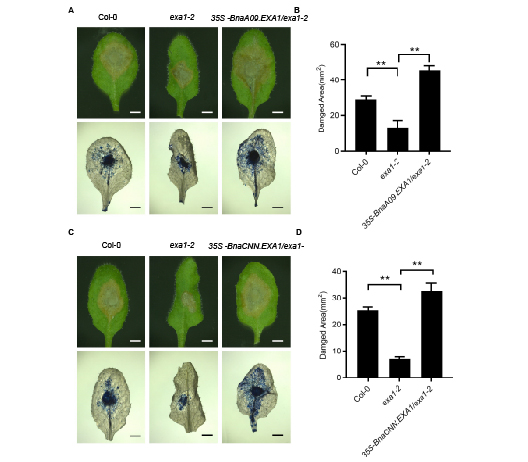
Loss function of EXA1 in Arabidopsis thaliana has been shown to increase PR gene expression and enhance resistance to pathogens. Here, we cloned BnaEXA1 in Brassica napus and ectopically overexpressed in A. thaliana exa1-2 mutant. We found that overexpressed BnaEXA1 not only restored the morphological defects of the exa1-2 mutant, but also reduced the expression of PR1 and PR2, resulting in the susceptibility to Sclerotinia sclerotiorum and oomycetes H.a. Noco2 in exa1-2 mutant, indicating that BnaEXA1 plays an important role in regulating the basic resistance of plants.
| [1] | 费维新, 李强生, 吴新杰, 侯树敏, 陈凤祥, 王文相, 胡宝成 (2002). 利用栽培措施控制油菜菌核病的研究. 中国油料作物学报 24(3), 47-49. |
| [2] | 王爱荣 (2006). 核盘菌与拟南芥互作的分子机制研究. 博士论文. 福州: 福建农林大学. pp. 1-106. |
| [3] | Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen ZH, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng QD, Rollins JA, Lebrun MH, Dickman M(2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7, e1002230. |
| [4] | Ash MR, Faelber K, Kosslick D, Albert GI, Roske Y, Kofler M, Schuemann M, Krause E, Freund C (2010). Conserved ?-hairpin recognition by the GYF domains of Smy2 and GIGYF2 in mRNA surveillance and vesicular transport complexes. Structure 18, 944-954. |
| [5] | Barbetti MJ, Banga SK, Fu TD, Li YC, Singh D, Liu SY, Ge XT, Banga SS (2014). Comparative genotype reactions to Sclerotinia sclerotiorum within breeding populations of Brassica napus and B. juncea from India and China. Euphytica 197, 47-59. |
| [6] | Dai FM, Xu T, Wolf GA, He ZH (2006). Physiological and molecular features of the pathosystem Arabidopsis thaliana L.-Sclerotinia sclerotiorum Libert. J Integr Plant Biol 48, 44-52. |
| [7] | Giovannone B, Tsiaras WG, de la Monte S, Klysik J, Lautier C, Karashchuk G, Goldwurm S, Smith RJ (2009). GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum Mol Genet 18, 4629-4639. |
| [8] | Guo XM, Stotz HU (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Microbe Interact 20, 1384-1395. |
| [9] | Hale VA, Guiney EL, Goldberg LY, Haduong JH, Kwartler CS, Scangos KW, Goutte C (2012). Notch signaling is antagonized by SAO-1, a novel GYF-domain protein that interacts with the E3 ubiquitin ligase SEL-10 in Caenorhabditis elegans. Genetics 190, 1043-1057. |
| [10] | Hashimoto M, Neriya Y, Keima T, Iwabuchi N, Koinuma H, Hagiwara-Komoda Y, Ishikawa K, Himeno M, Maejima K, Yamaji Y, Namba S (2016). EXA1, a GYF domain protein, is responsible for loss-of-susceptibility to Plantago asiatica mosaic virus in Arabidopsis thaliana. Plant J 88, 120-131. |
| [11] | Kofler M, Motzny K, Freund C (2005). GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol Cell Proteomics 4, 1797-1811. |
| [12] | Letunic I, Doerks T, Bork P (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257-D260. |
| [13] | Li S, Li X, Zhou YJ (2018). Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res 247, 15-20. |
| [14] | Matsui H, Nomura Y, Egusa M, Hamada T, Hyon GS, Kaminaka H, Watanabe Y, Ueda T, Trujillo M, Shirasu K, Nakagami H (2017). The GYF domain protein PSIG1 dampens the induction of cell death during plant-pathogen interactions. PLoS Genet 13, e1007037. |
| [15] | Rowe HC, Walley JW, Corwin J, Chan EKF, Dehesh K, Kliebenstein DJ (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog 6, e1000861. |
| [16] | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731-2739. |
| [17] | Wu ZS, Huang S, Zhang XB, Wu D, Xia ST, Li X (2017). Regulation of plant immune receptor accumulation through translational repression by a glycine-tyrosine-phenylalanine (GYF) domain protein. eLife 6, e23684. |
/
| 〈 |
|
〉 |