

收稿日期: 2021-07-09
录用日期: 2021-11-20
网络出版日期: 2021-11-17
基金资助
宁夏回族自治区农业特色优势产业育种专项(NXNYYZ20200101);宁夏回族自治区重点研发计划(2021BBF02002);宁夏回族自治区重点研发计划(重大)重点项目(2019BBF02022)
Molecular Identification of Tomato Spotted Wilt Virus on Tomato in Yinchuan
Received date: 2021-07-09
Accepted date: 2021-11-20
Online published: 2021-11-17
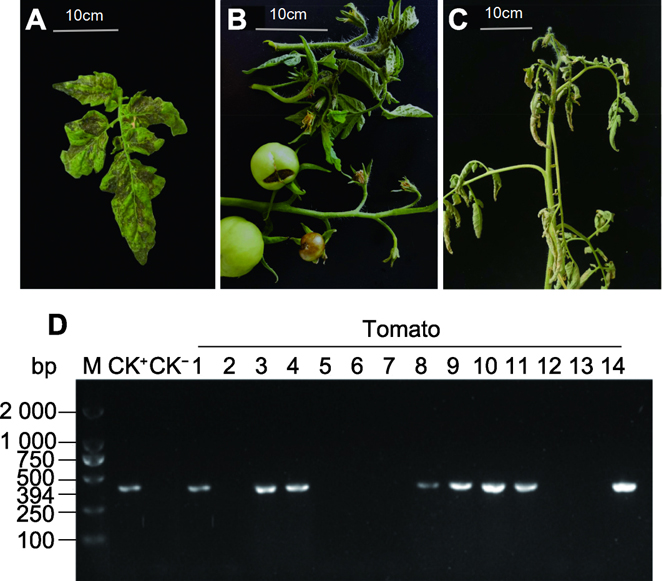
为明确银川番茄(Lycopersicon esculentum)是否遭受了番茄斑萎病毒(TSWV)的危害, 采用国家标准TSWV RT- PCR检测技术对银川番茄上采集的14份疑似感染TSWV病叶样本进行分子鉴定, 对克隆得到的核衣壳蛋白基因N (Nucleocapsid)序列进行多序列比对和系统进化树分析, 随后对PCR阳性样本进行蛋白检测。结果表明, 14份病叶样本中有8份扩增出长度为394 bp的TSWV N基因序列, 且8条序列完全一致; 获得的银川番茄TSWV分离物与云南番茄、中国莴苣(Lactuca sativa)、中国鸢尾(Iris tectorum)和重庆辣椒(Capsicum annuum) TSWV分离物相对近缘, 与山东、黑龙江和北京等地及国外TSWV分离物相对远缘; 利用TSWV的抗体通过Western blot对8个PCR阳性样本进一步检测, 结果也证实8个阳性样本中存在TSWV感染。该研究首次通过分子鉴定及蛋白检测证明银川番茄上存在TSWV感染, 需要加快抗TSWV番茄品种的选育工作。
关键词: 银川; 番茄斑萎病毒; RT-PCR; Western blot
王晓敏 , 李洪磊 , 王林 , 周鹏泽 , 白圣懿 , 李国花 , 郑福顺 , 陶小荣 , 程国新 , 高艳明 , 李建设 . 银川番茄斑萎病毒的分子鉴定[J]. 植物学报, 2021 , 56(6) : 715 -721 . DOI: 10.11983/CBB21113
In order to determine whether Yinchuan tomato (Lycopersicon esculentum) was damaged by tomato spotted wilt virus (TSWV), 14 disease leaf samples of suspected infected TSWV collected from Yinchuan tomato were identified by the national standard TSWV RT-PCR technique. The cloned N gene sequences were analyzed by multi sequence alignment and phylogenetic tree analysis. Eight of the 14 disease leaf samples amplified the TSWV N gene sequence with a length of 394 bp, and the 8 sequences were completely consistent. The TSWV sequence was relatively close to those from Yunnan tomato, Chinese lettuce (Lactuca sativa), Chinese Iris (Iris tectorum) and Chongqing pepper (Capsicum annuum), while relatively distant to those from Shandong, Heilongjiang, Beijing and the abroad. Detection of TSWV in the 8 PCR positive samples by use of Western bolt showed that TSWV infection existed in these samples. This study proved that there was TSWV infection in Yinchuan tomato. Therefore, it is necessary to speed up the breeding of TSWV resistant tomato varieties.

Key words: Yinchuan; tomato spotted wilt virus; RT-PCR; Western blot
| [1] | 陈东亮, 李明远, 李雪梅, 王玲玲, 黄丛林 (2018). 北京地区莴苣中番茄斑萎病毒的鉴定. 中国蔬菜 (1), 65-69. |
| [2] | 程国新 (2019). 辣椒(Capsicum annuum L.)可用叶色突变体的筛选鉴定及叶色调控基因CaPAL功能的初步分析. 博士论文. 杨凌: 西北农林科技大学. pp. 19-22. |
| [3] | 丁铭, 张丽珍, 方琦, 李婷婷, 苏晓霞, 李展, 张仲凯 (2004). 侵染马铃薯的一个Tospovirus混合分离物的鉴定、纯化及多抗血清制备. 西南农业学报 17(S1), 160-162. |
| [4] | 郭亚璐, 马晓飞, 史佳楠, 张柳, 张剑硕, 黄腾, 武鹏程, 康昊翔, 耿广荟, 陈浩, 魏健, 窦世娟, 李莉云, 尹长城, 刘国振 (2017). 转基因水稻中CAS9蛋白质的免疫印迹检测. 中国农业科学 50, 3631-3639. |
| [5] | 金凤媚, 宋建, 薛俊, 陈东亮, 王姝, 张越, 孙海波 (2020). 北京地区侵染菊花的番茄斑萎病毒分子检测与基因组部分序列分析. 华北农学报 35, 175-184. |
| [6] | 李艳莉, 温学萍, 徐苏萌, 杨俊丽, 汪洋, 张翔, 曹瑾 (2019). 宁夏越夏番茄产销现状分析报告. 园艺与种苗 39(6), 21-23. |
| [7] | 邱树亮, 王孝宣, 杜永臣, 高建昌, 国艳梅, 朱德蔚, 胡鸿, 李宝聚, 石延霞 (2012). 番茄斑萎病毒TSWV的鉴定及抗病种质的筛选. 园艺学报 39, 1107-1114. |
| [8] | 饶雪琴, 刘勇, 李媛媛, 吴竹妍 (2010). 广东番茄上检测到Tospovirus病毒. 植物病理学报 40, 430-432. |
| [9] | 孙淼, 荆陈沉, 楚成茹, 吴根土, 孙现超, 谢艳, 刘勇, 青玲 (2017). 重庆辣椒上番茄斑萎病毒的血清学检测及分子鉴定. 园艺学报 44, 487-494. |
| [10] | 王凯娜, 战斌慧, 周雪平 (2019). 黑龙江地区番茄斑萎病毒的鉴定及其部分生物学特征分析. 植物保护 45, 37-43. |
| [11] | 吴淑华, 涂丽琴, 咸文荣, 闫佳会, 高丹娜, 吉颖, 陶小荣, 周益军, 郭青云, 季英华 (2020). 青海辣椒上番茄斑萎病毒检测及鉴定. 园艺学报 47, 1391-1400. |
| [12] | 吴鑫本 (2008). 番茄斑萎病毒的防治与检疫. 现代农业科学 15(11), 86, 96. |
| [13] | 张万红, 冯佳, Ali K, 罗健达, 杨举田, 王术科, 宗浩, 申莉莉, 李莹, 王凤龙, 张石飞, 杨金广, 金轲 (2020). 山东烟区首次发现番茄斑萎病毒侵染. 中国烟草科学 41(5), 87-91. |
| [14] | 张友军, 吴青君, 徐宝云, 朱国仁 (2003). 危险性外来入侵生物--西花蓟马在北京发生危害. 植物保护 29(4), 58-59. |
| [15] | 中华人民共和国国家质量监督检验检疫总局, 中国国家标准化管理委员会 (2013). 番茄斑萎病毒PCR检测方法: GB/T 28982-2012. 北京: 中国标准出版社. |
| [16] | Adkins S (2000). Tomato spotted wilt virus-positive steps towards negative success. Mol Plant Pathol 1, 151-157. |
| [17] | Brittlebank CC (1919). Tomato diseases. J Agric 27, 213-235. |
| [18] | Burland TG (2000). DNASTAR's Lasergene sequence analysis software. Methods Mol Biol 132, 71-91. |
| [19] | Chiemsombat P, Adkins S (2006). Tospoviruses. In: Rao GP, Lava Kumar P, Holguín-Peña RJ, eds. Characteriza-tion, Diagnosis and Management of Plant Viruses. Hous-ton: Studium Press. pp. 1-37. |
| [20] | De Haan P, Kormelink R, De Oliveira Resende R, Van Poelwijk F, Peters D, Goldbach R (1991). Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol 72, 2207-2216. |
| [21] | De Haan P, Wagemakers L, Peters D, Goldbach R (1990). The S RNA segment of tomato spotted wilt virus has an ambisense character. J Gen Virol 71, 1001-1007. |
| [22] | Hayes S, Velanis CN, Jenkins GI, Franklin KA (2014). UV-B detected by the UVR-8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA 111, 11894-11899. |
| [23] | Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R (1992). The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambi-sense RNA segments. J Gen Virol 73, 2795-2804. |
| [24] | Mahmood T, Yang PC (2012). Western blot: technique, theory, and trouble shooting. North Am J Med Sci 4, 429-434. |
| [25] | Peiró A, Cañizares MC, Rubio L, López C, Moriones E, Aramburu J, Sánchez-Navarro J (2014). The movement protein (NSm) of tomato spotted wilt virus is the avirulen-ce determinant in the tomato Sw-5 gene-based resistan-ce. Mol Plant Pathol 15, 802-813. |
| [26] | Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD (2011). Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12, 938-954. |
| [27] | Soler S, Cebolla-Cornejo J, Nuez F (2003). Control of disease induced by tospoviruses in tomato: an update of the genetic approach. Phytopathol Med 42, 207-219. |
| [28] | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary dis-tance, and maximum parsimony methods. Mol Biol Evol 28, 2731-2739. |
/
| 〈 |
|
〉 |