

基于TurboID的植物蛋白邻近标记实验方法
收稿日期: 2021-07-01
录用日期: 2021-08-09
网络出版日期: 2021-08-11
基金资助
国家自然科学基金(31870171);国家自然科学基金(31701246)
Methods for TurboID-based Proximal Labeling in Plants
Received date: 2021-07-01
Accepted date: 2021-08-09
Online published: 2021-08-11
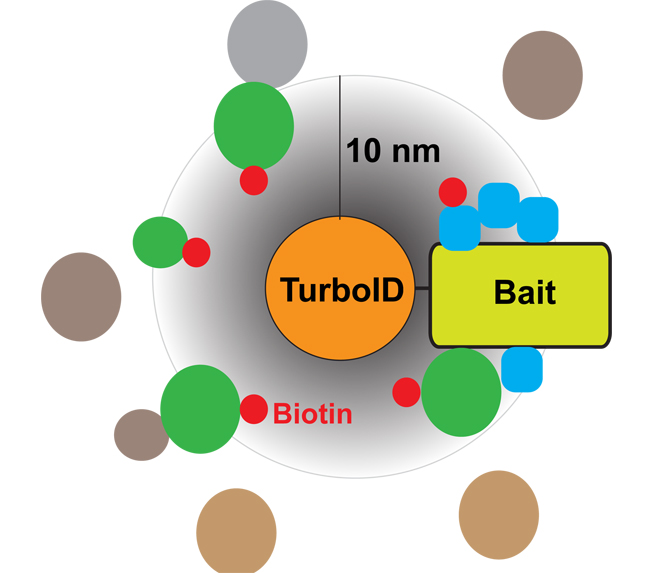
邻近标记作为近些年发展起来的一项检测活细胞内蛋白互作关系和亚细胞结构蛋白组的新型技术, 已成功应用于多种动植物体系的研究。该技术通过给诱饵蛋白融合一个具有特定催化连接活性的酶, 在酶的催化作用下将小分子底物(如生物素)共价连接到酶邻近的内源蛋白, 通过富集和分析被标记的蛋白可获得与诱饵互作的蛋白组。经定向进化产生的生物素连接酶TurboID具有无蛋白毒性及催化效率高的优势。利用TurboID介导的邻近标记技术分析感兴趣蛋白的邻近蛋白组, 可研究细胞内瞬时发生或微弱的蛋白互作网络, 进而解析复杂的生物学过程。该文详细描述了在拟南芥(Arabidopsis thaliana)中基于TurboID的邻近标记实验方法及注意事项, 旨在为利用这一新技术研究植物蛋白互作关系提供参考。
邝嘉怡, 李洪清, 沈文锦, 高彩吉 . 基于TurboID的植物蛋白邻近标记实验方法[J]. 植物学报, 2021 , 56(5) : 584 -593 . DOI: 10.11983/CBB21104
Proximity labeling (PL), a recently developed technique to detect protein-protein interactions and subcellular structural proteomes in living cells, has been successfully applied in various animal and plant systems. Proximity labeling is conducted by fusing an engineered enzyme with catalytic activity to a protein of interest (bait protein). With the catalysis of the enzyme, small molecular substrates such as biotin are covalently linked to endogenous proximal proteins, which can be further enriched and analyzed to identify the interactome of the bait protein. TurboID, a biotin ligase produced by directed evolution, has the advantages of non-toxicity and high catalytic efficiency. Using TurboID-based proximity labeling to analyze proximal proteins of bait proteins, we can study transient or weak protein interactions, which helps to understand the complex biological processes occurring inside cells. Here, we describe methods and related tips for TurboID-based proximal labeling in Arabidopsis thaliana, and hope to provide a reference for studying plant protein-protein interactions.

Key words: TurboID; proximity labeling; biotin; protein interactions
| [1] | 李喜豹, 赖敏怡, 梁山, 王小菁, 高彩吉, 杨超 (2021). 植物细胞自噬基因的功能与转录调控机制. 植物学报 56, 201-217. |
| [2] | 刘洋, 张静, 王秋玲, 侯岁稳 (2018). 植物细胞自噬研究进展. 植物学报 53, 5-16. |
| [3] | 苏田, 韩笑, 刘华东 (2020). 邻近标记在蛋白质组学中的发展及应用. 中国生物化学与分子生物学报 36, 36-41. |
| [4] | Alban C, Job D, Douce R (2000). Biotin metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol 51, 17-47. |
| [5] | Arora D, Abel NB, Liu C, Van Damme P, Yperman K, Eeckhout D, Vu LD, Wang J, Tornkvist A, Impens F, Korbei B, Van Leene J, Goossens A, De Jaeger G, Ott T, Moschou PN, Van Damme D (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32, 3388-3407. |
| [6] | Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880-887. |
| [7] | Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee HW, Lee IK, Carr SA, Ting AY (2020). Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci USA 117, 12143-12154. |
| [8] | Kim TW, Park CH, Hsu CC, Zhu JY, Hsiao Y, Branon T, Xu SL, Ting AY, Wang ZY (2019). Application of TurboID mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. bioRxiv doi: https://doi.org/10.1101/636324. |
| [9] | Lin QP, Zhou ZJ, Luo WB, Fang MC, Li MR, Li HQ (2017). Screening of proximal and interacting proteins in rice protoplasts by proximity-dependent biotinylation. Front Plant Sci 8, 749. |
| [10] | Mair A, Xu SL, Branon TC, Ting AY, Bergmann DC (2019). Proximity labeling of protein complexes and cell- type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8, e47864. |
| [11] | Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30, 1143-1148. |
| [12] | Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801-810. |
| [13] | Yang C, Shen WJ, Yang LM, Sun Y, Li XB, Lai MY, Wei J, Wang CJ, Xu YC, Li FQ, Liang S, Yang CW, Zhong SW, Luo M, Gao CJ (2020). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant 13, 515-531. |
| [14] | Yang XX, Wen ZY, Zhang DL, Li Z, Li DW, Nagalakshmi U, Dinesh-Kumar SP, Zhang YL (2021). Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Commun 2, 100137. |
| [15] | Zhang YL, Song GY, Lal NK, Nagalakshmi U, Li YY, Zheng WJ, Huang PJ, Branon TC, Ting AY, Walley JW, Dinesh-Kumar SP (2019). TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun 10, 3252. |
/
| 〈 |
|
〉 |