

甜蜜的相遇—营养与激素信号协同调节植物生长的新机制
收稿日期: 2021-02-24
录用日期: 2021-02-26
网络出版日期: 2021-03-03
基金资助
国家自然科学基金(31970308);国家自然科学基金重大研究计划(91740203)
A Sweet Meet—New Mechanism on Nutrient and Hormone Regulation of Plant Growth
Received date: 2021-02-24
Accepted date: 2021-02-26
Online published: 2021-03-03
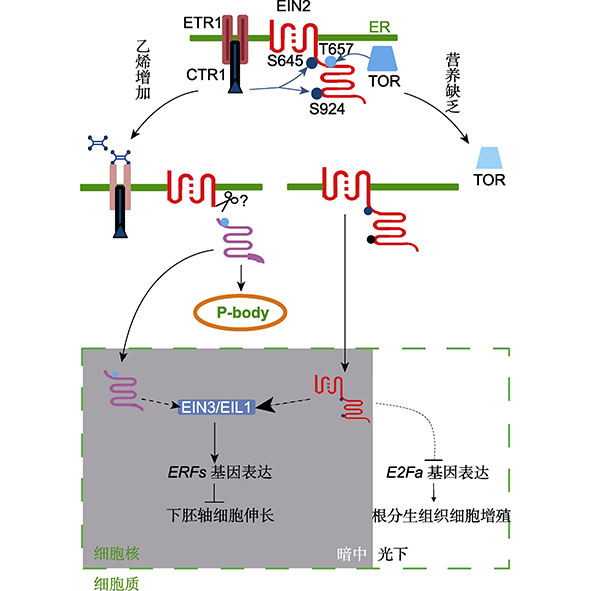
为应对持续不断的环境压力和逆境胁迫, 植物需要整合内部和外部信息来调整自身的生长发育, 以适应环境。其中, 可溶性糖不仅是基础能量和营养代谢的必需分子, 也是参与植物生长发育和应对胁迫的信号分子。然而, 植物整合糖信号, 平衡营养代谢和胁迫应答的分子机制尚不清楚。最近, 福建农林大学熊延团队发现, 居于植物营养感受通路中心地位的TOR激酶能够直接磷酸化乙烯信号核心组分EIN2蛋白, 形成1个葡萄糖-TOR-EIN2的营养感受和调控轴心。植物通过不同的蛋白激酶(TOR和CTR1)精确调控EIN2不同位点的磷酸化, 从而使EIN2成为葡萄糖信号和乙烯信号的交叉中心, 精巧地调节植物的生长发育。
温兴 , 晋莲 , 郭红卫 . 甜蜜的相遇—营养与激素信号协同调节植物生长的新机制[J]. 植物学报, 2021 , 56(2) : 138 -141 . DOI: 10.11983/CBB21040
Continuously exposed to a variety of environmental stresses, plants need to integrate internal and external information in order to achieve the purpose of adaption to environment. Among this process the perception and regulation of the energy state and soluble sugar level are of great importance. However, the molecular mechanisms underlying the integration of sugar signaling, nutrient metabolism and stress response in plants remain unclear. Recently, a team led by Prof. Yan Xiong from Fujian Agriculture and Forestry University (FAFU) have discovered that TOR kinase, the central player in metabolic signaling pathways, can directly bind and phosphorylate the core component of ethylene signaling EIN2 protein, forming a regulatory axis to coordinate TOR and ethylene signaling. The two kinases TOR and CTR1 precisely regulates distinct phosphorylation sites on EIN2, respectively, which makes EIN2 become a coordination hub of glucose signal and ethylene signal, and precisely control plant growth and development.

Key words: glucose; ethylene; TOR; EIN2; phosphorylation
| [1] | Chen RQ, Binder BM, Garrett WM, Tucker ML, Chang C, Cooper B (2011). Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol Biosyst 7,2637-2650. |
| [2] | Depaepe T, Hendrix S, van Rensburg HCJ Van den Ende W, Cuypers A, Van Der Straeten D (2021). At the crossroads of survival and death: the reactive oxygen species-ethylene-sugar triad and the unfolded protein response. Trends Plant Sci 26,338-351. |
| [3] | Fu LW, Liu YL, Qin GC, Wu P, Zi HL, Xu ZT, Zhao XD, Wang Y, Li YX, Yang SH, Peng C, Wong CCL, Yoo SD, Zuo ZC, Liu RY, Cho YH, Xiong Y (2021). The TOR- EIN2 axis mediates nuclear signaling to modulate plant growth. Nature 591,288-292. |
| [4] | Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, B?uerlein FJB, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grünewald K (2015). Structural basis of vesicle formation at the inner nuclear membrane. Cell 163,1692-1701. |
| [5] | Hao DD, Sun XZ, Ma B, Zhang JS, Guo HW (2017). Ethylene. In: Li JY, Li CY, Smith SM, eds. Hormone Metabolism and Signaling in Plants. London: Academic Press. pp.203-241. |
| [6] | Ingargiola C, Duarte GT, Robaglia C, Leprince AS, Meyer C (2020). The plant target of rapamycin: a conductor of nutrition and metabolism in photosynthetic organisms. Genes (Basel) 11, 1285. |
| [7] | Ju CL, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, Chang C, (2012). CTR1 phos- phorylates the central regulator EIN 2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109, 19486-19491. |
| [8] | Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC (2007). Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci USA 104,7241-7246. |
| [9] | Li WY, Ma MD, Feng Y, Li HJ, Wang YC, Ma YT, Li MZ, An FY, Guo HW (2015). EIN2-directed translational regu- lation of ethylene signaling in Arabidopsis. Cell 163, 670- 683. |
| [10] | Pandey BK, Huang GQ, Bhosale R, Hartman S, Sturrock CJ, Jose L, Martin OC, Martin M, Voesenek LACJ, Ljung K, Lynch JP, Brown KM, Whalley WR, Mooney SJ, Zhang DB, Bennett MJ (2021). Plant roots sense soil compaction through restricted ethylene diffusion. Science 371,276-280. |
| [11] | Qiao H, Shen ZX, Huang SSC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethy- lene gas. Science 338,390-393. |
| [12] | Shen X, Li YL, Pan Y, Zhong SW (2016). Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front Plant Sci 7,1571. |
| [13] | Wang PC, Zhao Y, Li ZP, Hsu CC, Liu X, Fu LW, Hou YJ, Du YY, Xie SJ, Zhang CG, Gao JH, Cao MJ, Huang XS, Zhu YF, Tang K, Wang XG, Tao WA, Xiong Y, Zhu JK (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69,100-112. |
| [14] | Wen X, Zhang CL, Ji YS, Zhao Q, He WR, An FY, Jiang LW, Guo HW (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22,1613-1616. |
| [15] | Wu Y, Shi L, Li LW, Fu LW, Liu YL, Xiong Y, Sheen J (2019). Integration of nutrient, energy, light, and hormone signaling via TOR in plants. J Exp Bot 70,2227-2238. |
| [16] | Xiong Y, McCormack M, Li L, Hall Q, Xiang CB, Sheen J (2013). Glucose-TOR signaling reprograms the transcriptome and activates meristems. Nature 496,181-186. |
| [17] | Yuan XB, Xu P, Yu YD, Xiong Y (2020). Glucose-TOR signaling regulates PIN2 stability to orchestrate auxin gradient and cell expansion in Arabidopsis root. Proc Natl Acad Sci USA 117,32223-32225. |
| [18] | Zhong SW, Shi H, Xue C, Wei N, Guo HW, Deng XW (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA 111,3913-3920. |
| [19] | Zhu FG, Deng J, Chen H, Liu P, Zheng LH, Ye QY, Li R, Brault M, Wen JQ, Frugier F, Dong JL, Wang T (2020). A cep peptide receptor-like kinase regulates auxin bio- synthesis and ethylene signaling to coordinate root growth and symbiotic nodulation in Medicago truncatula. Plant Cell 32,2855-2877. |
/
| 〈 |
|
〉 |