

基于CRISPR编辑系统的DNA片段删除技术
收稿日期: 2020-12-11
录用日期: 2021-02-25
网络出版日期: 2021-02-25
基金资助
广东省基础与应用基础研究重大项目(2019B030302006)
CRISPR-based DNA Fragment Deletion in Plants
Received date: 2020-12-11
Accepted date: 2021-02-25
Online published: 2021-02-25
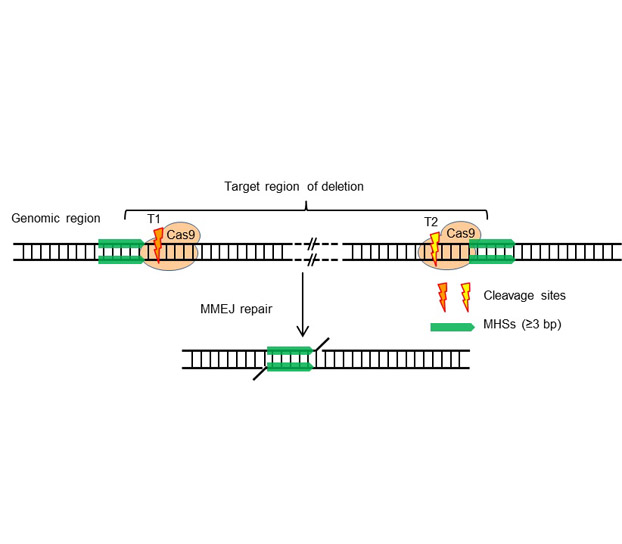
基于CRISPR/Cas9系统的基因组编辑技术已成为基因功能研究和遗传修饰的重要工具。在引导RNA的引导下, Cas9蛋白对基因组靶位点进行精准切割产生DNA双链断裂(DSB), 借助细胞内的DSB修复机制, 可实现基因组靶位点碱基的缺失、插入或者替换, 甚至发生片段删除。该文介绍了基于CRISPR/Cas9基因组编辑系统的DSB微同源末端连接修复方式(MMEJ)提高基因组DNA片段删除效率的方法, 主要从靶点设计和突变检测方面展开详细描述。
关键词: 基因组编辑; CRISPR/Cas9; MMEJ; 微同源; 片段删除
谢先荣 , 曾栋昌 , 谭健韬 , 祝钦泷 , 刘耀光 . 基于CRISPR编辑系统的DNA片段删除技术[J]. 植物学报, 2021 , 56(1) : 44 -49 . DOI: 10.11983/CBB20203
CRISPR/Cas9-based genome editing technology has been an important tool to study the gene function and genomic modification. Directed by a guide RNA, Cas9 protein can cleavage the genomic DNA at the target site, and produce mutations, including deletion, insertion, substitution and fragment deletion, by DNA double strand break (DSB) repair mechanism. In this protocol, we introduce the method to use CRISPR/Cas9 system to increase the efficiency of genomic DNA fragment deletion with microhomology-mediated end joining, especially the details in target design and detection of mutant plants.

Key words: genome editing; CRISPR/Cas9; MMEJ; microhomologous; fragment deletion
| [1] | 苏钺凯, 邱镜仁, 张晗, 宋振巧, 王建华 (2019). CRISPR/ Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报 54, 385-395. |
| [2] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光 (2018). 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法. 中国科学: 生命科学 48, 783-794. |
| [3] | Burgess DG, Xu J, Freeling M (2015). Advances in understanding cis regulation of the plant gene with an emphasis on comparative genomics. Curr Opin Plant Biol 27, 141-147. |
| [4] | Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH (2014). Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRIS- PR)/Cas9 nuclease system in mammalian cells. J Biol Chem 289, 21312-21324. |
| [5] | Liu WZ, Xie XR, Ma XL, Li J, Chen JH, Liu YG (2015). DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8, 1431-1433. |
| [6] | Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274-1284. |
| [7] | Ma XL, Zhu QL, Chen YL, Liu YG (2016). CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant 9, 961-974. |
| [8] | Manova V, Gruszka D (2015). DNA damage and repair in plants-from models to crops. Front Plant Sci 6, 885. |
| [9] | Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, Codner GF, Hublitz P, McHugh PJ, Teboul L, de Bruijn MFTR (2019). Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47, 7402-7417. |
| [10] | Tan JT, Zhao YC, Wang B, Hao Y, Wang YX, Li YY, Luo WN, Zong WB, Li GS, Chen SF, Ma K, Xie XR, Chen LT, Liu YG, Zhu QL (2020). Efficient CRISPR/Cas9-based plant genomic fragment deletions by microhomology- mediated end joining. Plant Biotechnol J 18, 2161-2163. |
| [11] | Tuladhar R, Yeu Y, Piazza JT, Tan ZP, Clemenceau JR, Wu XF, Barrett Q, Herbert J, Mathews DH, Kim J, Hwang TH, Lum L (2019). CRISPR-Cas9-based mu- tagenesis frequently provokes on-target mRNA misregulation. Nat Commun 10, 4056. |
| [12] | Xie XR, Liu WZ, Dong G, Zhu QH, Liu YG (2020). MMEJ-KO: a web tool for designing paired CRISPR guide RNAs for microhomology-mediated end joining fragment deletion. Sci China Life Sci doi: 10.1007/s11427-020-1797-3. |
| [13] | Xie XR, Ma XL, Zhu QL, Zeng DC, Li GS, Liu YG (2017). CRISPR-GE: a convenient software toolkit for CRISPR- based genome editing. Mol Plant 10, 1246-1249. |
| [14] | Zhang Y, Massel K, Godwin ID, Gao CX (2018). Applications and potential of genome editing in crop improvement. Genome Biol 19, 210. |
/
| 〈 |
|
〉 |