

甘蓝型油菜MADS-box基因家族的鉴定与系统进化分析
收稿日期: 2016-12-12
录用日期: 2017-05-22
网络出版日期: 2017-05-22
基金资助
西北农林科技大学博士科研启动资金(No.Z109021614)、基本科研业务专项资金(No.Z109021703)和唐仲英作物育种基金(No. A212021713)
Genome-wide Survey and Phylogenetic Analysis of MADS-box Gene Family in Brassica napus
Received date: 2016-12-12
Accepted date: 2017-05-22
Online published: 2017-05-22
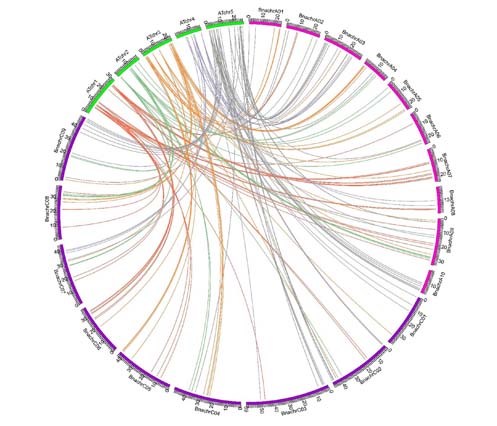
MADS-box基因家族参与调控开花时间、花器官分化、根系生长、分生组织分化、子房和配子发育、果实膨大及衰老等植物生长发育的重要过程。基于甘蓝型油菜(Brassica napus)基因组测序数据, 利用生物信息学方法对甘蓝型油菜MADS-box基因家族进行鉴定和注释及基因结构与系统进化分析。结果显示, 在甘蓝型油菜中鉴定出307个MADS-box基因家族成员, 根据进化关系可将其分为两大类型, I型(M-type)包含α、β、γ三个亚家族, II型(MIKC-type)包括MIKCC和MIKC*两个亚家族, MIKCC可进一步分为13个小类; 甘蓝型油菜A基因组染色体上分布的MADS-box基因多于C基因组。在基因结构上, MIKC-type亚家族基因序列普遍比M-type长且含有较多的外显子; M-type亚家族蛋白序列中的motif数量为2-5个, MIKC-type亚家族蛋白序列中平均含有7个motif。拟南芥(Arabidopsis thaliana)与甘蓝型油菜MADS-box基因共线性分析结果显示, 全基因组复制事件对MADS-box基因家族尤其是MIKC亚家族的扩张起重要作用; MIKC亚家族基因在进化过程中受到的选择压力约为M-type的2倍, 这表明MIKC-type亚家族在进化过程中被选择性保留。
高虎虎 , 张云霄 , 胡胜武 , 郭媛 . 甘蓝型油菜MADS-box基因家族的鉴定与系统进化分析[J]. 植物学报, 2017 , 52(6) : 699 -712 . DOI: 10.11983/CBB16244
The MADS-box gene family is involved in many processes during plant growth and development, such as flowering time, floral organ differentiation, root growth, meristem differentiation, ovary and gamete development, fruit enlargement and senescence. In this study, we used rape (Brassica napus) genome sequencing data with bioinformatics methods to identify and annotate the MADS-box genes. Rape contains 307 members of MADS-box gene family. According to the evolutionary relationships, these genes can be divided into two subfamilies: I-type, also known as M-type, containing three subclades, α, β, and γ; II type, also known as MIKC-type containing two subclades, MIKCC and MIKC*. MIKCC can be further divided into 13 groups. The number of MADS-box genes is greater in the A than C subgenome chromosome of B. napus. For the gene structure, the sequence is longer for MIKC-type than M-type genes and contains more exons. The number of motifs in M-type genes is about 2-5, and MIKC-type genes contain an average of 7 motifs. Synteny analysis revealed that whole-genome duplication played a major role in the expansion of the BnaMADS gene family, especially the MIKC-type subfamily. The selection pressure of the MIKC-type subfamily was about 2 times that for the M-type, which resulted in the selective preservation of MIKC-type subfamily genes during evolution.

Key words: Brassica napus; MADS-box; gene structure; evolution; synteny
| [1] | Airoldi CA, Davies B (2012). Gene duplication and the evolution of plant MADS-box transcription factors.J Genet Genomics 39, 157-165. |
| [2] | Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF (2000a). MADS-box gene evolution beyond flowers: expres- sion in pollen, endosperm, guard cells, roots and trichom- es.Plant J 24, 457-466. |
| [3] | Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Bur- geff C, Ditta GS, de Pouplana LR, Martínez-Castilla L, Yanofsky MF (2000b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals.Proc Natl Acad Sci USA 97, 5328-5333. |
| [4] | Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genomics 8, 242. |
| [5] | Bowers JE, Chapman BA, Rong JK, Paterson AH (2003). Unravelling angiosperm genome evolution by phylogene- tic analysis of chromosomal duplication events.Nature 422, 433-438. |
| [6] | Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014). Early allopolyploid evolution in the post- Neolithic Brassica napus oilseed genome. Science 345, 950-953. |
| [7] | Chang YY, Chiu YF, Wu JW, Yang CH (2009). Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol 50, 1425-1438. |
| [8] | Cheng F, Liu SY, Wu J, Fang L, Sun SL, Liu B, Li PX, Hua W, Wang XW (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11, 136. |
| [9] | Day RC, Herridge RP, Ambrose BA, Macknight RC (2008). Transcriptome analysis of proliferating Arabidopsis endos- perm reveals biological implications for the control of syn- cytial division, cytokinin signaling, and gene expression regulation.Plant Physiol 148, 1964-1984. |
| [10] | De Bodt S, Raes J, Van de Peer Y, Thei?en G (2003). And then there were many: MADS goes genomic.Trends Plant Sci 8, 475-483. |
| [11] | Díaz-Riquelme J, Lijavetzky D, Martínez-Zapater JM, Carmona MJ (2009). Genome-wide analysis of MIKCC- type MADS box genes in grapevine.Plant Physiol 149, 354-369. |
| [12] | Doebley J, Lukens L (1998). Transcriptional regulators and the evolution of plant form.Plant Cell 10, 1075-1082. |
| [13] | Duan WK, Song XM, Liu TK, Huang ZN, Ren J, Hou XL, Li Y (2015). Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol Genet Genomics 290, 239-255. |
| [14] | Edger PP, Pires JC (2009). Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes.Chromosome Res 17, 699-717. |
| [15] | Fan CM, Wang X, Wang YW, Hu RB, Zhang XM, Chen JX, Fu YF (2013). Genome-wide expression analysis of soy- bean MADS genes showing potential function in the seed development.PLoS One 8, e62288. |
| [16] | Fang SC, Fernandez DE (2002). Effect of regulated over- expression of the MADS domain factor AGL15 on flower senescence and fruit maturation.Plant Physiol 130, 78-89. |
| [17] | Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014). Pfam: the protein families database.Nucleic Acids Res 42, D222-D230. |
| [18] | Gan YB, Filleur S, Rahman A, Gotensparre S, Forde BG (2005). Nutritional regulation of ANR1 and other root- expressed MADS-box genes in Arabidopsis thaliana. Planta 222, 730-742. |
| [19] | Gramzow L, Ritz MS, Thei?en G (2010). On the origin of MADS-domain transcription factors.Trends Genet 26, 149-153. |
| [20] | Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals.Ann Bot 103, 1165-1172. |
| [21] | Grimplet J, Martínez-Zapater JM, Carmona MJ (2016). Structural and functional annotation of the MADS-box transcription factor family ingrapevine.BMC Genomics 17, 80. |
| [22] | Hemming MN, Trevaskis B (2011). Make hay when the sun shines: the role of MADS-box genes intemperature- dependant seasonal flowering responses.Plant Sci 180, 447-453. |
| [23] | Immink RGH, Kaufmann K, Angenent GC (2010). The ‘ABC’ of MADS domain protein behaviour and interactions.Semin Cell Dev Biol 21, 87-93. |
| [24] | Jin JP, Zhang H, Kong L, Gao G, Luo JC (2014). Plant TFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors.Nucleic Acids Res 42, D1182-D1187. |
| [25] | Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG, Childs KL, Davidson RM, Lin HN, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. |
| [26] | Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses.Mol Biol Evol 20, 1963-1977. |
| [27] | Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009). Circos: an information aesthetic for comparative genomics.Genome Res 19, 1639-1645. |
| [28] | Letunic I, Doerks T, Bork P (2015). SMART: recent upda- tes, new developments and status in 2015.Nucleic Acids Res 43, D257-D260. |
| [29] | Liu SY, Liu YM, Yang XH, Tong CB, Edwards D, Parkin IAP, Zhao MX, Ma JX, Yu JY, Huang SM, Wang XY, Wang JY, Lu K, Fang ZY, Bancroft I, Yang TJ, Hu Q, Wang XF, Yue Z, Li HJ, Yang LF, Wu J, Zhou Q, Wang WX, King GJ, Pires JC, Lu CX, Wu ZY, Sampath P, Wang Z, Guo H, Pan SK, Yang LM, Min JM, Zhang D, Jin DC, Li WS, Belcram H, Tu JX, Guan M, Qi CK, Du DZ, Li JN, Jiang LC, Batley J, Sharpe AG, Park BS, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong CH, Wang L, Li JP, Hu ZY, Zhuang M, Huang Y, Huang JY, Shi JQ, Mei DS, Liu J, Lee TH, Wang JP, Jin HZ, Li ZY, Li X, Zhang JF, Xiao L, Zhou YM, Liu ZS, Liu XQ, Qin R, Tang X, Liu WB, Wang YP, Zhang YY, Lee J, Kim HH, Denoeud F, Xu X, Liang XM, Hua W, Wang XW, Wang J, Chalhoub B, Paterson AH (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5, 3930. |
| [30] | Liu Y, Cui SJ, Wu F, Yan S, Lin XL, Du XQ, Chong K, Schilling S, Thei?en G, Meng Z (2013). Functional con- servation of MIKC*-type MADS box genes in Arabidopsis and rice pollen maturation.Plant Cell 25, 1288-1303. |
| [31] | Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005). Modeling gene and genome duplications in eukaryotes.Proc Natl Acad Sci USA 102, 5454-5459. |
| [32] | Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011). The emerging importance of type I MADS box transcription factors for plant reproduction.Plant Cell 23, 865-872. |
| [33] | Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJA, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang HZ, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi HY, Thomas PD, Finn RD (2015). The InterPro protein families data- base: the classification resource after 15 years.Nucleic Acids Res 43, D213-D221. |
| [34] | Nagaharu U (1935). Genome analysis in Brassica with spe- cial reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7, 389-452. |
| [35] | Nakano T, Suzuki K, Fujimura T, Shinshi H (2006). Genome- wide analysis of the ERF gene family in Arabidopsis and rice.Plant Physiol 140, 411-432. |
| [36] | Nam J, dePamphilis CW, Ma H, Nei M (2003). Antiquity and evolution of the MADS-box gene family controlling flower development in plants.Mol Biol Evol 20, 1435-1447. |
| [37] | Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003). Molecular and phylo- genetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world.Plant Cell 15, 1538-1551. |
| [38] | Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF (1995). Molecular evolution of flower development: diver- sification of the plant MADS-box regulatory gene family.Genetics 140, 345-356. |
| [39] | Saha G, Park JI, Jung HJ, Ahmed NU, Kayum MA, Chung MY, Hur Y, Cho YG, Watanabe M, Nou IS (2015). Genome-wide identification and characterization of MADS- box family genes related to organ development and stress resistance in Brassica rapa. BMC Genomics 16, 178. |
| [40] | Shao SQ, Li BY, Zhang ZT, Zhou Y, Jiang J, Li XB (2010). Expression of a cotton MADS-box gene is regulated in anther development and in response to phytohormone sig- naling.J Genet Genomics 37, 805-816. |
| [41] | Shore P, Sharrocks AD (1995). The MADS-box family of transcription factors.Eur J Biochem 229, 1-13. |
| [42] | Shu YJ, Yu DS, Wang D, Guo DL, Guo CH (2013). Genome- wide survey and expression analysis of the MADS-box gene family in soybean.Mol Biol Rep 40, 3901-3911. |
| [43] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: molecular evolutionary genetics analysis version 6.0.Mol Biol Evol 30, 2725-2729. |
| [44] | Tang HB, Bowers JE, Wang XY, Ming R, Alam M, Pater- son AH (2008). Synteny and collinearity in plant genomes.Science 320, 486-488. |
| [45] | Tapia-López R, García-Ponce B, Dubrovsky JG, Garay- Arroyo A, Pérez-Ruíz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008). An AGAMOUS-related MADS- box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146, 1182-1192. |
| [46] | The Brassica rapa Genome Sequencing Project Con- sortium, Wang XW, Wang HZ, Wang J, Sun RF, Wu J, Liu SY, Bai YQ, Mun JH, Bancroft I, Cheng F, Huang SW, Li XX, Hua W, Wang JY, Wang XY, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu BH, Li B, Liu B, Tong CB, Song C, Duran C, Peng CF, Geng CY, Koh C, Lin CY, Edwards D, Mu DS, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonn- ema G, Tang HB, Wang HP, Belcram H, Zhou HL, Hirakawa H, Abe H, Guo H, Wang H, Jin HZ, Parkin IAP, Batley J, Kim JS, Just J, Li JW, Xu JH, Deng J, Kim JA, Li JP, Yu JY, Meng JL, Wang JP, Min JM, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao MX, Jin MN, Ramchiary N, Drou N, Berkman PJ, Cai QL, Huang QF, Li RQ, Tabata S, Cheng SF, Zhang S, Zhang SJ, Huang SM, Sato SS, Sun SL, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li YR, Du YC, Liao YC, Lim Y, Narusaka Y, Wang YP, Wang ZY, Li ZY, Wang ZW, Xiong ZY, Zhang ZH (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43, 1035-1039. |
| [47] | Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H (2000). A short history of MADS-box genes in plants.Plant Mol Biol 42, 115-149. |
| [48] | Tiwari S, Spielman M, Schulz R, Oakey RJ, Kelsey G, Salazar A, Zhang K, Pennell R, Scott RJ (2010). Trans- criptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol 10, 72. |
| [49] | Wei B, Zhang RZ, Guo JJ, Liu DM, Li AL, Fan RC, Mao L, Zhang XQ (2014). Genome-wide analysis of the MADS- box gene family in Brachypodium distachyon. PLoS One 9, e84781. |
| [50] | Wei X, Wang LH, Yu JY, Zhang YX, Li DH, Zhang XR (2015). Genome-wide identification and analysis of the MADS-box gene family insesame.Gene 569, 66-76. |
| [51] | Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang XW (2014). Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids.Proc Natl Acad Sci USA 111, 5283-5288. |
| [52] | Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyse- linck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010). Arabidopsis female gametophyte gene expression map reveals similarities bet- ween plant and animal gametes.Curr Biol 20, 506-512. |
| [53] | Xu ZD, Zhang QX, Sun LD, Du DL, Cheng TR, Pan HT, Yang WR, Wang J (2014). Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Mol Genet Genomics 289, 903-920. |
| [54] | Yang JH, Liu DY, Wang XW, Ji CM, Cheng F, Liu BN, Hu ZY, Chen S, Pental D, Ju YH, Yao P, Li XM, Xie K, Zhang JH, Wang JL, Liu F, Ma WW, Shopan J, Zheng HK, Mackenzie SA, Zhang MF (2016). The genome se- quence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selec- tion. Nat Genet 48, 1225-1232. |
| [55] | Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles trans- cription factors. Nature 346, 35-39. |
| [56] | Yao QY, Xia EH, Liu FH, Gao LZ (2015). Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 557, 35-42. |
| [57] | Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, Xiang CB (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals.Mol Plant 7, 1653-1669. |
| [58] | Zhang Z, Li J, Zhao XQ, Wang J, Wong GKS, Yu J (2006). KaKs_calculator: calculating Ka and Ks through model selection and model averaging.Genomics Proteomics Bioinformatics 4, 259-263. |
/
| 〈 |
|
〉 |