

药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达
? 共同第一作者
收稿日期: 2014-08-07
录用日期: 2015-01-05
网络出版日期: 2015-09-06
基金资助
山东省自然科学基金(No;ZR2012CM024)和中国科学院系统与进化国家重点实验室开放课题(No.LSEB2011-01)
Sequence Divergence and Expression Specificity of the Starch Synthase Gene Family in Oryza officinalis Leaf
? These authors contributed equally to this paper
Received date: 2014-08-07
Accepted date: 2015-01-05
Online published: 2015-09-06
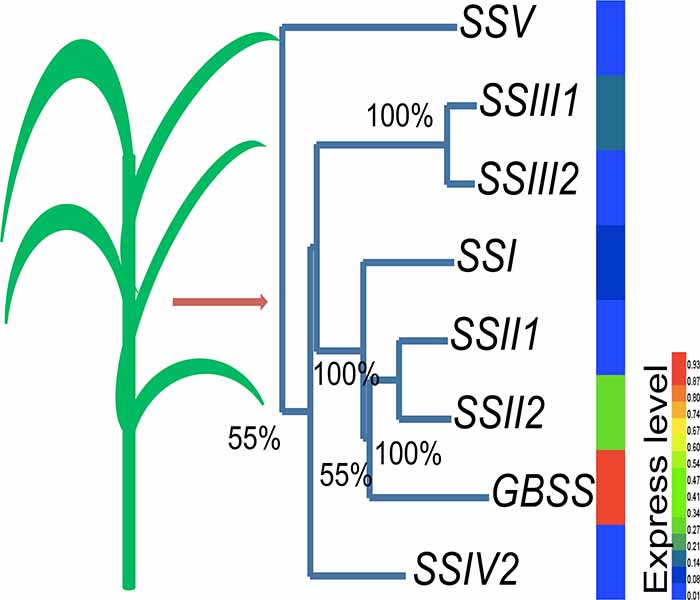
淀粉不仅是植物自身和后代生长繁殖的重要营养与能量储备, 而且是人类膳食中碳水化合物的主要来源。植物中淀粉合成主要发生在两个阶段, 一是在形成临时淀粉的光合作用阶段, 另一个则是在成为贮藏淀粉的营养积累阶段。相对于最后的淀粉贮藏阶段, 临时淀粉的形成阶段在植物整个碳水化合物代谢过程中扮演着更为重要的角色, 然而却一直少有关注。为深入研究初始淀粉合成过程中相关酶在植物中的进化模式, 选取了药用野生稻(Oryza officinalis)为研究对象, 通过对其全叶转录组的重测序, 定性、定量地调查了淀粉合成酶基因家族在稻属野生物种光合器官中的基因类型和表达变化。共有8个淀粉合成酶基因的完整编码序列在药用野生稻的叶中首次被识别。系统发育分析表明, 这8个基因分别隶属SSI、SSII、SSIII、SSIV、SSV和GBSSII基因家族。序列比较和相对表达定量分析显示, 药用野生稻与栽培稻的淀粉合成酶基因家族的进化模式具有高度的一致性, 两个物种的同源基因在mRNA水平的序列相似度达到95%-98%。基于非同义置换和同义置换比率的统计检验表明, 8个基因在两个物种间均经历了严格的纯化选择。另外, 3个在栽培稻胚乳中特异表达的基因在药用野生稻的叶转录组中未筛查出来, 而4个在栽培稻叶中优势表达的基因在药用野生稻叶中同样呈现相对较高水平的表达。
景翔 , 包颖 , 杜家潇 , 徐思 . 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达[J]. 植物学报, 2015 , 50(6) : 683 -690 . DOI: 10.11983/CBB14147
Starch is produced by most green plants as an energy store and is also a major carbohydrate source in human diet. There are two stages of starch synthesis in most plants. In the first stage, transient starch is produced during photosynthesis, and in the second, the nutrition accumulation stage, starch is deposited for storage purposes. Although the first stage of starch synthesis plays a critical role in carbohydrate metabolism, starch biosynthesis in photosynthetic organs has received less attention. To detect the evolutionary patterns associated with starch synthase, the enzyme involved in the biosynthesis of starch, in plant photosynthetic organs, we used leaf transcriptome resequencing to investigate the genotype and expression divergence of the gene family in Oryza officinalis. Eight complete starch synthase genes were identified, and phylogenetic analyses confirmed that the genes were SSI, SSII, SIII, SSIV, SSV, and GSBBII. Comparison of sequences and expression quantities showed that the evolutionary patterns of the starch synthase family in O. officinalis were highly consistent with those of cultivated rice. The homologous sequence identities between the two species were 95% to 98% at the mRNA level. Further statistical tests based on the ratio of nonsynonymous to synonymous substitutions indicated that all eight genes were under significant purifying selection. In addition, three genes specifically expressed in the endosperm of cultivated rice were not found in the leaf transcriptome of O. officinalis. However, four genes with predominant expression in the leaves of cultivated rice showed relatively higher expression in the O. officinalis leaf transcriptome.

| [1] | Ammiraju JSS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, Talag J, Fan CZ, Goicoechea JL, Zuccolo A, Chen JF, Bennetzen JL, Chen MS, Jackson S, Wing RA (2008). Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set.Plant Cell 20, 3191-3209. |
| [2] | Brust H, Lehmann T, D'Hulst C, Fettke J (2014). Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin.PLoS One 9, e102364. |
| [3] | Buléon A, Colonna P, Planchot V, Ball S (1998). Starch granules: structure and biosynthesis.Int J Biol Macromol 23, 85-112. |
| [4] | Cao HP, Imparl-Radosevich J, Guan HP, Keeling PL, James MG, Myers AM (1999). Identification of the soluble starch synthase activities of maize endosperm.Plant Phy- siol 120, 205-216. |
| [5] | Deschamps P, Moreau H, Worden AZ, Dauvillée D, Ball SG (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts.Genetics 178, 2373-2387. |
| [6] | Dian WM, Jiang HW, Wu P (2005). Evolution and expression analysis of starch synthase III and IV in rice.J Exp Bot 56, 623-632. |
| [7] | Fujita N (2014). Starch biosynthesis in rice endosperm.AGri-Biosci Monogr 4, 1-18. |
| [8] | Fujita N, Satoh R, Hayashi A, Kodama M, Itoh R, Aihara S, Nakamura Y (2011). Starch biosynthesis in rice endosp- erm requires the presence of either starch synthase I or IIIa.J Exp Bot 62, 4819-4831. |
| [9] | Gámez-Arjona FM, Li J, Raynaud S, Baroja-Fernández E, Munñoz FJ, Ovecka M, Ragel P, Bahaji A, Pozueta-Romero J, Mérida Á (2011). Enhancing the expression of starch synthase class IV results in increased levels of both transitory and long-term storage starch.Plant Biotechnol J 9, 1049-1060. |
| [10] | Garber M, Grabherr MG, Guttman M, Trapnell C (2011). Computational methods for transcriptome annotation and quantification using RNA-seq.Nat Methods 8, 469-477. |
| [11] | Goldman N, Yang ZH (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences.Mol Biol Evol 11, 725-736. |
| [12] | Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224. |
| [13] | Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.Syst Biol 59, 307-321. |
| [14] | Hirose T, Terao T (2004). A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.).Planta 220, 9-16. |
| [15] | Jiang HW, Dian WM, Liu FY, Wu P (2004). Molecular cloning and expression analysis of three genes encoding starch synthase II in rice.Planta 218, 1062-1070. |
| [16] | Langmead B, Salzberg SL (2012). Fast gapped-read align- ment with Bowtie 2.Nat Methods 9, 357-359. |
| [17] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools.Bioinformatics 25, 2078-2079. |
| [18] | Lu F, Ammiraju JSS, Sanyal A, Zhang SL, Song RT, Chen JF, Li GS, Sui Y, Song X, Cheng ZK, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen MS (2009). Comparative sequence analysis of MONOCULM1-orthol- ogous regions in 14 Oryza genomes.Proc Natl Acad Sci USA 106, 2071-2076. |
| [19] | Myers AM, Morell MK, James MG, Ball SG (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal.Plant Physiol 122, 989-998. |
| [20] | Nei M, Gojobori T (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions.Mol Biol Evol 3, 418-426. |
| [21] | Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [22] | Orzechowski S (2008). Starch metabolism in leaves.Acta Biochim Pol 55, 435-445. |
| [23] | Schwarte S, Brust H, Steup M, Tiedemann R (2013). Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana.BMC Res Notes 6, 84. |
| [24] | Stamova BS, Laudencia-Chingcuanco D, Beckles DM (2009). Transcriptomic analysis of starch biosynthesis in the developing grain of hexaploid wheat.Int J Plant Genomics 2009, 407426. |
| [25] | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation.Nat Biotechnol 28, 511-515. |
| [26] | Yang ZH, Nielsen R (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models.Mol Biol Evol 17, 32-43. |
| [27] | Zeeman SC, Kossmann J, Smith AM (2010). Starch: its metabolism, evolution, and biotechnological modification in plants.Annu Rev Plant Biol 61, 209-234. |
| [28] | Zeeman SC, Smith SM, Smith AM (2007). The diurnal metabolism of leaf starch.Biochem J 401, 13-28. |
| [29] | Zhang XL, Szydlowski N, Delvallé D, D'Hulst C, James MG, Myers AM (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis.BMC Plant Biol 8, 96. |
/
| 〈 |
|
〉 |