

光受体介导信号转导调控植物开花研究进展
收稿日期: 2018-06-25
录用日期: 2018-09-17
网络出版日期: 2019-07-31
基金资助
国家自然科学基金(31471907, No.31530064)
Advances in Photoreceptor-mediated Signaling Transduction in Flowering Time Regulation
Received date: 2018-06-25
Accepted date: 2018-09-17
Online published: 2019-07-31
马朝峰 , 戴思兰 . 光受体介导信号转导调控植物开花研究进展[J]. 植物学报, 2019 , 54(1) : 9 -22 . DOI: 10.11983/CBB18147
Light is an important environmental factor that affects plant growth and development. Flowering is the most important event in higher plants. Plants perceive accurately changes in the surrounding light environments by photoreceptors, thus activating a series of signaling transduction processes and initiating flowering. Here, we summarized the current understanding of the structural characteristics and physiological functions of various photoreceptors in higher plants. We reviewed the molecular mechanisms of phytochromes, cryptochromes, and FKF1/ZTL/LKP2 in mediating signaling transduction and flowering time, including transcriptional and post-transcriptional regulation of CO and FT. Finally, we described the advances in photoreceptor-mediated-integration of light, temperature, and gibberellin signals in regulating flowering. Future directions in this area were also proposed.
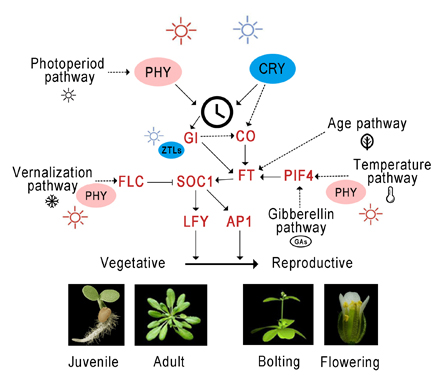
Key words: photoreceptor; flowering; phytochrome; cryptochrome
| 1 | 许淑娟, 种康 ( 2018). “先驱”转录因子LEC1在早期胚胎重置春化状态的机制. 植物学报 53, 1-4. |
| 2 | Ahmad M, Cashmore AR ( 1993). HY4 gene of A.thaliana encodes a protein with characteristics of a blue-light photoreceptor..Nature 366, 162-166. |
| 3 | Amasino R ( 2010). Seasonal and developmental timing of flowering. Plant J 61, 1001-1013. |
| 4 | Andrés F, Coupland G ( 2012). The genetic basis of flowe- ring responses to seasonal cues. Nat Rev Genet 13, 627-639. |
| 5 | Bae G, Choi G ( 2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59, 281-311. |
| 6 | Balasubramanian S, Sureshkumar S, Lempe J, Weigel D ( 2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature.PLoS Genet 2, e106. |
| 7 | Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T ( 2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606-622. |
| 8 | Bendix C, Marshall CM, Harmon FG ( 2015). Circadian clock genes universally control key agricultural traits. Mol Plant 8, 1135-1152. |
| 9 | Blázquez MA, Ahn JH, Weigel D ( 2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana.Nat Genet 33, 168-171. |
| 10 | Blümel M, Dally N, Jung C ( 2015). Flowering time regulation in crops-what did we learn from Arabidopsis? Curr Opin Biotechnol 32, 121-129. |
| 11 | Briggs WR, Christie JM ( 2002). Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7, 204-210. |
| 12 | Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED ( 2003). Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell 11, 59-67. |
| 13 | Cai XN, Ballif J, Endo S, Davis E, Liang MX, Chen D, DeWald D, Kreps J, Zhu T, Wu YJ ( 2007). A putative CCAAT-binding transcription factor is a regulator of flowe- ring timing in Arabidopsis. Plant Physiol 145, 98-105. |
| 14 | Cao SH, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF ( 2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis.Plant Cell 26, 1009-1017. |
| 15 | Cashmore AR ( 2003). Cryptochromes: enabling plants and animals to determine circadian time. Cell 114, 537-543. |
| 16 | Cashmore AR, Jarillo JA, Wu YJ, Liu DM ( 1999). Cryptochromes: blue light receptors for plants and animals. Science 284, 760-765. |
| 17 | Castillejo C, Pelaz S ( 2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering.Curr Biol 18, 1338-1343. |
| 18 | Celaya RB, Liscum E ( 2005). Phototropins and associated signaling: providing the power of movement in higher plants. Photochem Photobiol 81, 73-80. |
| 19 | Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M ( 2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62, 335-364. |
| 20 | Chen M, Chory J ( 2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21, 664-671. |
| 21 | Chen XM ( 2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development.Science 303, 2022-2025. |
| 22 | Christie JM ( 2007). Phototropin blue-light receptors. Annu Rev Plant Biol 58, 21-45. |
| 23 | Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O'Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, Jenkins GI, Getzoff ED ( 2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335, 1492-1496. |
| 24 | Coelho CP, Minow MAA, Chalfun-Júnior A, Colasanti J ( 2014). Putative sugarcane FT/TFL1 genes delay flowe- ring time and alter reproductive architecture in Arabidopsis.Front Plant Sci 5, 221. |
| 25 | Corbesier L, Vincent C, Jang S, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G ( 2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030-1033. |
| 26 | de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S ( 2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480-484. |
| 27 | de Lucas M, Prat S ( 2014). PIFs get BRright: phytochrome interacting factors as integrators of light and hormonal signals. New Phytol 202, 1126-1141. |
| 28 | Devlin PF, Kay SA ( 2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499-2510. |
| 29 | Dotto M, Gómez MS, Soto MS, Casati P ( 2018). UV-B radiation delays flowering time through changes in the PRC2 complex activity and miR156 levels in Arabidopsis tha-liana.Plant Cell Environ 41, 1394-1406. |
| 30 | Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A ( 2013). PHYTOCHROME-DEPENDENT LATE-FLOWE- RING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci USA 110, 18017-18022. |
| 31 | Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, Seidlitz HK, Nagy F, Ulm R ( 2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28, 591-601. |
| 32 | Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G ( 2009). Arabidopsis DOF trans- cription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response.Dev Cell 17, 75-86. |
| 33 | Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G, Mizoguchi T ( 2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowe- ring in Arabidopsis. Plant Cell 20, 2960-2971. |
| 34 | Galv?o VC, Horrer D, Küttner F, Schmid M ( 2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana.Development 139, 4072-4082. |
| 35 | Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R ( 2010). Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 107, 20132-20137. |
| 36 | Guo HW, Yang HY, Mockler TC, Lin CT ( 1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360-1363. |
| 37 | Halliday KJ, Salter MG, Thingnaes E, Whitelam GC ( 2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33, 875-885. |
| 38 | Helliwell CA, Wood CC, Robertson M, Peacock WJ, Dennis ES ( 2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex.Plant J 46, 183-192. |
| 39 | Heschel MS, Selby J, Butler C, Whitelam GC, Sharrock RA, Donohue K ( 2007). A new role for phytochromes in temperature-dependent germination. New Phytol 174, 735-741. |
| 40 | Huq E, Tepperman JM, Quail PH ( 2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Ara- bidopsis. Proc Natl Acad Sci USA 97, 9789-9794. |
| 41 | Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA ( 2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis.Science 309, 293-297. |
| 42 | I?igo SS, Alvarez MJ, Strasser B, Califano A, Cerdán PD ( 2012). PFT1, the MED25 subunit of the plant mediator complex, promotes flowering through CONSTANS depen- dent and independent mechanisms in Arabidopsis. Plant J 69, 601-612. |
| 43 | Ito S, Song YH, Imaizumi T ( 2012 a). LOV domain-contai- ning F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant 5, 573-582. |
| 44 | Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T ( 2012 b). Flowering BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis . Proc Natl Acad Sci USA 109, 3582-3587. |
| 45 | Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G ( 2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27, 1277-1288. |
| 46 | Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR ( 2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487-490. |
| 47 | Jenkins GI ( 2014). The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26, 21-37. |
| 48 | Johansson M, Staiger D ( 2014). SRR1 is essential to repress flowering in non-inductive conditions in Arabidopsis thaliana.J Exp Bot 65, 5811-5822. |
| 49 | Johansson M, Staiger D ( 2015). Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot 66, 719-730. |
| 50 | Johnson E, Bradley M, Harberd NP, Whitelam GC ( 1994). Photoresponses of light-grown phyA mutants of Arabidopsis (Phytochrome A is required for the perception of daylength extensions).Plant Physiol 105, 141-149. |
| 51 | Kami C, Lorrain S, Hornitschek P, Fankhauser C ( 2010). Chapter two—light-regulated plant growth and development. Curr Top Dev Biol 91, 29-66. |
| 52 | Kim DH, Doyle MR, Sung SB, Amasino RM ( 2009). Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25, 277-299. |
| 53 | Kim J, Geng RS, Gallenstein RA, Somers DE ( 2013). The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140, 4060-4069. |
| 54 | Koornneef M, Hanhart CJ, van der Veen JH ( 1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana.Mol Gen Genet 229, 57-66. |
| 55 | Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA ( 2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242-245. |
| 56 | Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ ( 2008). The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long- day photoperiods in Arabidopsis. Planta 228, 709-723. |
| 57 | Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U ( 2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213-3222. |
| 58 | Lazaro A, Valverde F, Pi?eiro M, Jarillo JA ( 2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24, 982-999. |
| 59 | Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH ( 2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis.Genes Dev 21, 397-402. |
| 60 | Leivar P, Monte E ( 2014). PIFs: systems integrators in plant development. Plant Cell 26, 56-78. |
| 61 | Li F, Sun JJ, Wang DH, Bai SN, Clarke AK, Holm M ( 2014). The B-Box Family Gene STO (BBX24) in Arabidopsis tha- liana regulates flowering time in different pathways.PLoS One 9, e87544. |
| 62 | Liang T, Mei SL, Shi C, Yang Y, Peng Y, Ma LB, Wang F, Li X, Huang X, Yin YH, Liu HT ( 2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44, 512-523. |
| 63 | Liu B, Zuo ZC, Liu HT, Liu XM, Lin CT ( 2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25, 1029-1034. |
| 64 | Liu HT, Wang Q, Liu YW, Zhao XY, Imaizumi T, Somers DE, Tobin EM, Lin CT ( 2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA 110, 17582-17587. |
| 65 | Liu HT, Yu XH, Li KW, Klejnot J, Yang HY, Lisiero D, Lin CT ( 2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535-1539. |
| 66 | Mao J, Zhang YC, Sang Y, Li QH, Yang HQ ( 2005). From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102, 12270-12275. |
| 67 | Más P, Kim WY, Somers DE, Kay SA ( 2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana.Nature 426, 567-570. |
| 68 | Matsushita T, Mochizuki N, Nagatani A ( 2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571-574. |
| 69 | Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G ( 2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis.Plant Cell 17, 2255-2270. |
| 70 | Monte E, Alonso JM, Ecker JR, Zhang YL, Li X, Young J, Austin-Phillips S, Quail PH ( 2003). Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways.Plant Cell 15, 1962-1980. |
| 71 | Morris K, Thornber S, Codrai L, Richardson C, Craig A, Sadanandom A, Thomas B, Jackson S ( 2010). DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days.Plant Cell 22, 1118-1128. |
| 72 | Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B ( 2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331-340. |
| 73 | Osnato M, Castillejo C, Matías-Hernández L, Pelaz S ( 2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3, 808. |
| 74 | Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RGH, Schmid M ( 2013). Temperature- dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414-417. |
| 75 | Qian CZ, Mao WW, Liu Y, Ren H, Lau OS, Ouyang XH, Huang X ( 2016). Dual-source nuclear monomers of UV-B light receptor direct photomorphogenesis in Arabidopsis. Mol Plant 9, 1671-1674. |
| 76 | Rataj K, Simpson GG ( 2014). Message ends: RNA 3' pro- cessing and flowering time control. J Exp Bot 65, 353-363. |
| 77 | Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP ( 2002). MicroRNAs in plants. Genes Dev 16, 1616-1626. |
| 78 | Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Sch?fer E, Nagy F, Jenkins GI, Ulm R ( 2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103-106. |
| 79 | Rockwell NC, Su YS, Lagarias JC ( 2006). Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57, 837-858. |
| 80 | Salathia N, Davis SJ, Lynn JR, Michaels SD, Amasino RM, Millar AJ ( 2006). FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol 6, 10. |
| 81 | Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz- Sommer Z, Yanofsky MF, Coupland G ( 2000). Distinct roles of CONSTANS target genes in reproductive deve- lopment of Arabidopsis. Science 288, 1613-1616. |
| 82 | Samach A, Wigge PA ( 2005). Ambient temperature perception in plants. Curr Opin Plant Biol 8, 483-486. |
| 83 | Sawa M, Kay SA ( 2011). GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana.Proc Natl Acad Sci USA 108, 11698-11703. |
| 84 | Sawa M, Nusinow DA, Kay SA, Imaizumi T ( 2007). FKF1 and GIGANTEA complex formation is required for day- length measurement in Arabidopsis. Science 318, 261-265. |
| 85 | Sch?fer E, Bowler C ( 2002). Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep 3, 1042-1048. |
| 86 | Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA ( 2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13, 2659-2670. |
| 87 | Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A ( 2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189-201. |
| 88 | Somers DE, Kim WY, Geng RS ( 2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16, 769-782. |
| 89 | Somers DE, Schultz TF, Milnamow M, Kay SA ( 2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis . Cell 101, 319-329. |
| 90 | Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T ( 2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045-1049. |
| 91 | Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C ( 2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function.Genes Dev 17, 256-268. |
| 92 | Steinbach Y, Hennig L ( 2014). Arabidopsis MSI1 functions in photoperiodic flowering time control. Front Plant Sci 5, 77. |
| 93 | Strasser B, Alvarez MJ, Califano A, Cerdán PD ( 2009). A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J 58, 629-640. |
| 94 | Suetsugu N, Wada M ( 2013). Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol 54, 8-23. |
| 95 | Takano M, Inagaki N, Xie XZ, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T ( 2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17, 3311-3325. |
| 96 | Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T ( 2011). LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowe- ring under non-inductive conditions, dependent on FLAVIN- BINDING KELCH REPEAT F-BOX1.Plant J 67, 608-621. |
| 97 | Tao Z, Shen LS, Gu XF, Wang YZ, Yu H, He YH ( 2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124-128. |
| 98 | Teotia S, Tang GL ( 2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant 8, 359-377. |
| 99 | Tepperman JM, Hwang YS, Quail PH ( 2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48, 728-742. |
| 100 | Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ ( 2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-ele- ment.New Phytol 187, 57-66. |
| 101 | Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G ( 2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003-1006. |
| 102 | Wang CQ, Guthrie C, Sarmast MK, Dehesh K ( 2014). BBX19 interacts with CONSTANS to repress FLOWE- RING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis.Plant Cell 26, 3589-3602. |
| 103 | Wang FF, Lian HL, Kang CY, Yang HQ ( 2010). Phytochrome B is involved in mediating red light-induced sto- matal opening in Arabidopsis thaliana.Mol Plant 3, 246-259. |
| 104 | Wang JW ( 2014). Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65, 4723-4730. |
| 105 | Wang Q, Zuo ZC, Wang X, Gu LF, Yoshizumi T, Yang ZH, Yang L, Liu Q, Liu W, Han YJ, Kim JI, Liu B, Wohlschlegel JA, Matsui M, Oka Y, Lin CT ( 2016). Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354, 343-347. |
| 106 | Wu D, Hu Q, Yan Z, Chen W, Yan CY, Huang X, Zhang J, Yang PY, Deng HT, Wang JW, Deng XW, Shi YG ( 2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214-219. |
| 107 | Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS ( 2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750-759. |
| 108 | Xing SP, Salinas M, H?hmann S, Berndtgen R, Huijser P ( 2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935-3950. |
| 109 | Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T ( 2005). TWIN SISTER OF FT ( TSF) acts as a floral pathway integrator redundantly with FT.Plant Cell Physiol 46, 1175-1189. |
| 110 | Yan ZQ, Liang DW, Liu H, Zheng GC ( 2010). FLC: a key regulator of flowering time in Arabidopsis. Russ J Plant Physiol 57, 166-174. |
| 111 | Yang HQ, Wu YJ, Tang RH, Liu DM, Liu Y, Cashmore AR ( 2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815-827. |
| 112 | Yang LW, Fu JX, Qi S, Hong Y, Huang H, Dai SL ( 2017). Molecular cloning and function analysis of ClCRY1a and ClCRY1b, two genes in Chrysanthemum lavandulifolium that play vital roles in promoting floral transition.Gene 617, 32-43. |
| 113 | Yang Y, Liang T, Zhang LB, Shao K, Gu XX, Shang RX, Shi N, Li X, Zhang P, Liu HT ( 2018). UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants 4, 98-107. |
| 114 | Yasui Y, Kohchi T ( 2014). VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 repress the FLOWERING LOCUS C clade members to control flowering time in Arabidopsis.Biosci Biotechnol Biochem 78, 1850-1855. |
| 115 | Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T ( 2012). The phytochrome-interac-ting VASCULAR PLANT ONE-ZINC FINGER 1 and VOZ2 redundantly regulate flowering in Arabidopsis.Plant Cell 24, 3248-3263. |
| 116 | Yeom M, Kim H, Lim J, Shin AY, Hong S, Kim JI, Nam HG ( 2014). How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol Plant 7, 1701-1704. |
| 117 | Yu JW, Rubio V, Lee NY, Bai SL, Lee SY, Kim SS, Liu LJ, Zhang YY, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW ( 2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regula- ting GI stability. Mol Cell 32, 617-630. |
| 118 | Yuan S, Zhang ZW, Zheng C, Zhao ZY, Wang Y, Feng LY, Niu GQ, Wang CQ, Wang JH, Feng H, Xu F, Bao F, Hu Y, Cao Y, Ma LG, Wang HY, Kong DD, Xiao W, Lin HH, He YK ( 2016 a). Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc Natl Acad Sci USA 113, 7661-7666. |
| 119 | Yuan WY, Luo X, Li ZC, Yang WN, Wang YZ, Liu R, Du JM, He YH ( 2016b). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis.Nat Genet 48, 1527-1534. |
| 120 | Zuo ZC, Liu HT, Liu B, Liu XM, Lin CT ( 2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21, 841-847. |
/
| 〈 |
|
〉 |