

拟南芥组蛋白分子伴侣AtHIRA参与体细胞同源重组及盐胁迫响应
收稿日期: 2017-03-22
录用日期: 2017-08-30
网络出版日期: 2017-08-30
基金资助
国家自然科学基金(No.31671263)
Histone Chaperone AtHIRA is Involved in Somatic Homologous Recombination and Salinity Response in Arabidopsis
Received date: 2017-03-22
Accepted date: 2017-08-30
Online published: 2017-08-30
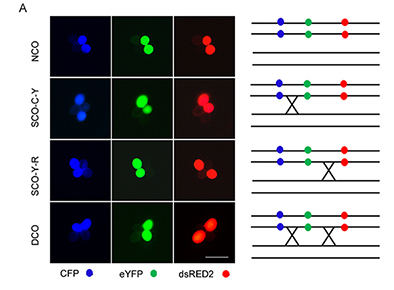
HIRA是组蛋白H3.3的特异分子伴侣, 在组蛋白H3.3掺入染色质的过程中发挥重要作用。研究表明, HIRA在哺乳动物胚胎发育和DNA损伤修复过程中不可或缺。而目前人们对于植物中HIRA同源基因功能的研究相对较少。该研究主要关注拟南芥(Arabidopsis thaliana) AtHIRA基因在植物体细胞同源重组以及减数分裂同源重组过程中的功能。将体细胞同源重组和减数分裂同源重组报告系统分别导入野生型和hira-1突变体后统计同源重组频率, 结果表明在正常生长条件下及在伯莱霉素(bleomycin)或UV-C处理条件下, hira-1突变体体细胞的分子内和分子间同源重组频率均低于野生型。而在正常生长条件下, 野生型与hira-1突变体花粉母细胞间的减数分裂同源重组频率没有明显差异, hira-1突变体的DNA损伤水平与野生型接近。qRT-PCR结果表明, DNA损伤修复相关基因RAD51和RAD54在hira-1突变体中的表达水平均高于野生型。此外, 盐胁迫处理实验表明, hira-1突变体对于高盐胁迫更加敏感。综上, AtHIRA在拟南芥体细胞同源重组及盐胁迫响应过程中发挥了一定作用。
陈成 , 董爱武 , 苏伟 . 拟南芥组蛋白分子伴侣AtHIRA参与体细胞同源重组及盐胁迫响应[J]. 植物学报, 2018 , 53(1) : 42 -50 . DOI: 10.11983/CBB17058
Histone regulator (HIRA) is a specific chaperone for histone H3.3 and plays an important role in the incorporation of histone H3.3 into chromatin. HIRA is indispensable in mammalian embryo development and DNA damage repair process, but we have few studies of the function of an HIRA homolog in plants. Here, we studied the function of Arabidopsis thaliana AtHIRA in somatic homologous recombination (HR) and meiotic homologous recombination. We used the somatic HR system and the meiotic homologous recombination system in wild type and the hira-1 mutant, a loss- of-function mutant of AtHIRA. Both intramolecular and intermolecular HR frequency was lower in hira-1 than the wild type under normal growth conditions and under bleomycin or UV-C treatment, with no significant difference in the frequency of meiotic recombination of microsporocytes between the wild type and hira-1 mutant under normal growth conditions. As well, under normal growth conditions or bleomycin treatment, loss-of-function of AtHIRA in Arabidopsis did not affect the DNA damage level. On qRT-PCR, the expression of RAD51 and RAD54, two DNA repair-related genes, was higher in hira-1 than the wild type. In addition, hira-1 had a salt-sensitive phenotype as compared with the wild type under salt stress. AtHIRA may play a role in somatic HR and the salinity response in Arabidopsis.

Key words: HIRA; homologous recombination; DNA damage repair; meiotic; salt stress
| [1] | 夏志强, 何奕昆, 鲍时来, 种康 (2007). 植物开花的组蛋白甲基化调控分子机理. 植物学通报 24, 275-283. |
| [2] | Amin AD, Vishnoi N, Prochasson P (2012). A global requirement for the HIR complex in the assembly of chromatin.Biochim Biophys Acta 1819, 264-276. |
| [3] | Andersen SL, Sekelsky J (2010). Meiotic versus mitotic recombination: two different routes for double-strand b- reak repair: the different functions of meiotic versus mi- totic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32, 1058-1066. |
| [4] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP (2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [5] | Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell 16, 479-485. |
| [6] | Gao J, Zhu Y, Zhou WB, Molinier J, Dong AW, Shen WH (2012). NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis.Plant Cell 24, 1437-1447. |
| [7] | Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI (2001). Homologous recombination in planta is stimulated in the absence of Rad50.EMBO Rep 2, 287-291. |
| [8] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [9] | Heyer WD, Ehmsen KT, Liu J (2010). Regulation of homologous recombination in eukaryotes.Annu Rev Genet 44, 113-139. |
| [10] | Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010). Zygotic resetting of the HISTONE 3 variant re- pertoire participates in epigenetic reprogramming in Arabi- dopsis.Curr Biol 20, 2137-2143. |
| [11] | Krejci L, Altmannova V, Spirek M, Zhao XL (2012). Homologous recombination and its regulation.Nucleic Acids Res 40, 5795-5818. |
| [12] | Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P (2005). The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus.Nature 437, 1386-1390. |
| [13] | Melamed-Bessudo C, Levy AA (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchro- matic but not in heterochromatic regions in Arabidopsis.Proc Natl Acad Sci USA 109, E981-E988. |
| [14] | Molinier J, Ries G, Bonhoeffer S, Hohn B (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16, 342-352. |
| [15] | Nie X, Wang HF, Li J, Holec S, Berger F (2014). The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics.Biol Open 3, 794-802. |
| [16] | Okada T, Endo M, Singh MB, Bhalla PL (2005). Analysis of the histoneH3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44, 557-568. |
| [17] | Otero S, Desvoyes B, Gutierrez C (2014). Histone H3 dynamics in plant cell cycle and development.Cytogenet Genome Res 143, 114-124. |
| [18] | Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ (2002). Targeted mutagenesis of theHira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryo- nic lethality. Mol Cell Biol 22, 2318-2328. |
| [19] | Schuermann D, Fritsch O, Lucht JM, Hohn B (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase δ mutants.Plant Cell 21, 2700-2714. |
| [20] | Schuermann D, Molinier J, Fritsch O, Hohn B (2005). The dual nature of homologous recombination in plants.Tren- ds Genet 21, 172-181. |
| [21] | Sherwood PW, Osley MA (1991). Histone regulatory (hir) mutations suppress δ insertion alleles in Saccharomyces cerevisiae. Genetics 128, 729-738. |
| [22] | Szenker E, Lacoste N, Almouzni G (2012). A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep 1, 730-740. |
| [23] | Tang Y, Poustovoitov MV, Zhao KH, Garfinkel M, Canu- tescu A, Dunbrack R, Adams PD, Marmorstein R (2006). Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly.Nat Struct Mol Biol 13, 921-929. |
| [24] | Wijeratne AJ, Ma H (2007). Genetic analyses of meiotic recombination in Arabidopsis. J Integr Plant Biol 49, 1199-1207. |
| [25] | Xu Y, Price BD (2011). Chromatin dynamics and the repair of DNA double strand breaks.Cell Cycle 10, 261-267. |
| [26] | Zhu Y, Dong AW, Meyer D, Pichon O, Renou JP, Cao KW, Shen WH (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18, 2879-2892. |
/
| 〈 |
|
〉 |