

利用PCR方法鉴定hiTAIL-PCR扩增产物中的质粒骨架片段
收稿日期: 2016-11-11
录用日期: 2017-04-17
网络出版日期: 2017-04-17
基金资助
国家自然科学基金(No.31200234)和陕西省教育厅省级重点实验室科研计划(No.14JS089)
PCR Used to Find Plasmid Backbone Fragments in the Products of hiTAIL-PCR
Received date: 2016-11-11
Accepted date: 2017-04-17
Online published: 2017-04-17
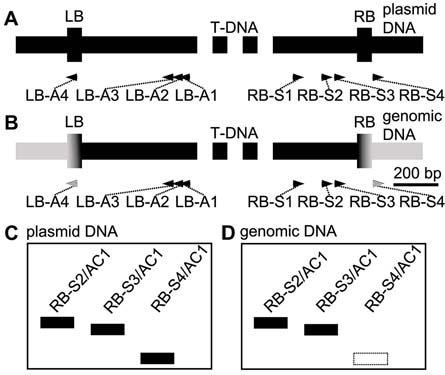
T-DNA突变体是研究基因功能的重要资源。高效热不对称交错PCR (hiTAIL-PCR)是克隆突变体中T-DNA插入位点侧翼序列的常用方法。然而我们发现, 利用hiTAIL-PCR克隆到的一些侧翼序列并不对应于宿主的染色体DNA序列, 而是质粒的骨架DNA片段。通过设置1组RB-S4/AC1或者LB-A4/AC1对照反应, 用PCR方法鉴定了hiTAIL-PCR扩增产物中位于T-DNA侧翼的质粒骨架片段。在后续分析中, 通过排除这些片段, 提高了利用hiTAIL-PCR获得宿主染色体DNA片段的效率。同时, 通过调整反应程序, 使得整个PCR的反应时间也大为缩短。在拟南芥(Arabidopsis thaliana) T-DNA突变体drf1侧翼序列的克隆实例中, 对照反应的引入将hiTAIL-PCR中需鉴定的22条扩增产物降至4条, 效率提高了81.8%。
关键词: hiTAIL-PCR; T-DNA突变体; 侧翼序列; 质粒骨架; 克隆
曹媛 , 杨云 , 徐化全 , 刘洋 , 王丹阳 . 利用PCR方法鉴定hiTAIL-PCR扩增产物中的质粒骨架片段[J]. 植物学报, 2018 , 53(1) : 104 -109 . DOI: 10.11983/CBB16216
T-DNA mutants are important resources for research of gene function. High-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) is widely used for cloning the flanking sequence near the T-DNA insertion sites. However, we found that some cloned flanking fragments in hiTAIL-PCR products corresponded not to the host genomic DNA but to the plasmid backbone DNA. In this study, with a control of the RB-S4/AC1 or LB-A4/AC1 product, we amplified PCR fragments from the plasmid backbone DNA. By excluding them from further analysis, we amplified fragments from the unknown genomic DNA more effectively. Meanwhile, by adjusting the PCR programs, the whole PCR time was greatly shortened. In cloning the flanking sequence of Arabidopsis thaliana T-DNA mutant drf1, our method with hiTAIL-PCR reduced the total 22 DNA bands required for further checking to 4 bands, which improved the efficiency by 81.8%.

Key words: hiTAIL-PCR; T-DNA mutant; flanking sequence; plasmid backbone; cloning
| [1] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012). Genome sequencing reveals agronomically important loci in rice using MutMap.Nat Biotechnol 30, 174-178. |
| [2] | Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657. |
| [3] | Coulondre C, Miller JH (1977). Genetic studies of the lac repressor: III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol 117, 525-567. |
| [4] | Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42, 819-832. |
| [5] | Jander G (2006). Gene identification and cloning by molecular marker mapping.Methods Mol Biol 323, 115-126. |
| [6] | Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002). Arabidopsis map-based cloning in the post-genome era.Plant Physiol 129, 440-450. |
| [7] | Kleinboelting N, Huep G, Appelhagen I, Viehoever P, Li Y, Weisshaar B (2015). The structural features of thousands of T-DNA insertion sites are consistent with a double- strand break repair-based insertion mechanism.Mol Plant 8, 1651-1664. |
| [8] | Liu YG, Chen YL (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences.Biotechniques 43, 649-650. |
| [9] | Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8, 457-463. |
| [10] | Martineau B, Voelker TA, Sanders RA (1994). On defining T-DNA.Plant Cell 6, 1032-1033. |
| [11] | Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991). T-DNA integration: a mode of illegitimate recombination in plants.EMBO J 10, 697-704. |
| [12] | Mueller PR, Wold B (1989). In vivo footprinting of a muscle specific enhancer by ligation mediated PCR.Science 246, 780-786. |
| [13] | Nan GL, Walbot V (2009). Plasmid rescue: recovery of flanking genomic sequences from transgenic transposon insertion sites.Methods Mol Biol 526, 101-109. |
| [14] | Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF (1990). A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones.Nucleic Acids Res 18, 2887-2890. |
| [15] | Stachel SE, Timmerman B, Zambryski P (1987). Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5' virD gene products. EMBO J 6, 857-863. |
| [16] | Triglia T, Peterson MG, Kemp DJ (1988). A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16, 8186. |
| [17] | Tzfira T, Li JX, Lacroix B, Citovsky V (2004). Agrobacterium T-DNA integration: molecules and models.Trends Ge- net 20, 375-383. |
| [18] | Wang HR, Fang J, Liang CZ, He MH, Li QY, Chu CC (2011). Computation-assisted SiteFinding-PCR for isolating flanking sequence tags in rice.Biotechniques 51, 421-423. |
| [19] | Zhou ZW, Ma HY, Qu LJ, Xie F, Ma QW, Ren ZR (2012). Establishment of an improved high-efficiency thermal asym- metric interlaced PCR for identification of genomic integration sites mediated by phiC31 integrase.World J Microbiol Biotechnol 28, 1295-1299. |
/
| 〈 |
|
〉 |