

利用低拷贝核基因重建菊科紫菀亚科族间系统发育关系
收稿日期: 2015-08-15
录用日期: 2015-09-05
网络出版日期: 2015-10-09
基金资助
国家自然科学基金(No.91131007)
Phylogenetic Reconstruction of Tribal Relationships in Asteroi- deae (Asteraceae) with Low-copy Nuclear Genes
Received date: 2015-08-15
Accepted date: 2015-09-05
Online published: 2015-10-09
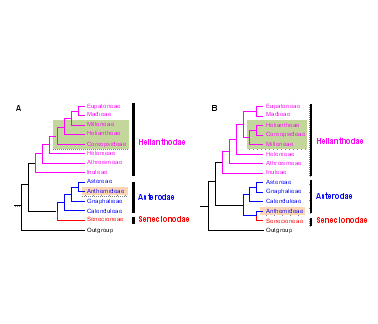
紫菀亚科(Asteroideae)是菊科最大的一个亚科, 包含的种数多于被子植物的绝大多数科。目前, 紫菀亚科族间的系统发育关系主要依赖于叶绿体基因信息, 但是叶绿体基因为单亲遗传, 并不能完整反映进化历史。鉴于杂交现象在菊科普遍存在, 故利用核基因可以反映更完整的紫菀亚科进化历史。该研究首次使用从转录组数据(20个新测+11个从NCBI数据库下载)中筛选出的47个直系同源低拷贝核基因来研究紫菀亚科的系统发育关系, 共选取了29个物种, 代表了紫菀亚科20个族中的13个族。用超矩阵分析方法和溯祖推测分析方法各获得了1个稳定的紫菀亚科系统树, 每个树上绝大多数分支都得到了高度支持, 且2个树之间没有明显的冲突。新的紫菀亚科族间系统发育关系揭示了千里光超族应并入紫菀超族, 春黄菊族可能是千里光族与紫菀族杂交起源的, 金鸡菊族很可能也是杂交起源的。该研究结果显示低拷贝核基因可以更好地解决科以下分类阶元的系统发育关系, 对菊科乃至被子植物其它科的系统发育研究具有重要的借鉴意义。
黄建勋 , 刘勉 , 张彩飞 , 马红 . 利用低拷贝核基因重建菊科紫菀亚科族间系统发育关系[J]. 植物学报, 2015 , 50(5) : 549 -564 . DOI: 10.11983/CBB15164
Asteroideae is the largest subfamily of the daisy family, Asteraceae, with more species than most other angiosperm families. Previous phylogenetic relationships of Asteroideae tribes were mostly based on chloroplast genes, which are uniparentally inherited and could not reflect the entire evolutionary histories. Especially considering the widespread phenomenon of hybridization in Asteraceae, low-copy nuclear genes with biparental inheritance can better serve as phylogenetic markers, for a more complete understanding of their evolution. To resolve the intertribal relationships in Asteroideae, transcriptomic datasets were newly generated by sequencing 20 species and additional datasets for 11 species downloaded from NCBI, representing 13 tribes and all 3 supertribes of this subfamily. Extensive sequence analyses of the 31 datasets resulted in the selection of 47 orthologous nuclear genes for phylogenetic reconstruction. Using the supermatrix analyses and coalescent estimates, we obtained robust and highly supported species trees, which are in agreement with each other. New intertribal phylogenies of Asteroideae revealed that: (1) the supertribe Senecionodae should be merged into the supertribe Asterodae; (2) Anthemideae possibly originated from a hybrid of ancestors of Senecioneae and Astereae; and (3) Coreopsideae is probably also hybrid originated. This study demonstrates that low-copy nuclear genes are good markers for infra-familial phylogenetic reconstruction and provides a foundation for future studies of phylogeny of Asteraceae and other angiosperm families.

Key words: Asteraceae; Asteroideae; low-copy nuclear genes; phylogeny; transcriptome-sequencing
| 1 | 刘勉 (2015). 用低拷贝核基因重建菊科植物的系统发育关系. 硕士论文. 上海: 复旦大学. |
| 2 | Baldwin BG (2009). Heliantheae alliance. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 689-711. |
| 3 | Baldwin BG, Wessa BL, Panero JL (2002). Nuclear rDNA evidence for major lineages of helenioid Heliantheae (Com- positae).Syst Bot 27, 161-198. |
| 4 | Bayer RJ, Starr JR (1998). Tribal phylogeny of the Asteraceae based on two non-coding chloroplast sequ- ences, the trnL intron and trnL/trnF intergenic spacer.Ann Mo Bot Gard 85, 242-256. |
| 5 | Bremer K (1987). Tribal interrelationships of the Aster- aceae.Cladistics 3, 210-253. |
| 6 | Bremer K, Anderberg AA (1994). Asteraceae: Cladistics & Classification. Portland: Timber Press. |
| 7 | Calabria LM, Emerenciano VP, Ferreira MJ, SCotti MT, Mabry TJ (2007). A phylogenetic analysis of tribes of the Asteraceae based on phytochemical data. Nat Prod Com- mun 2, 277-285. |
| 8 | Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses.Bioinformatics 25, 1972-1973. |
| 9 | Corriveau JL, Coleman AW (1988). Rapid screening me- thod to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species.Am J Bot 75, 1443-1458. |
| 10 | Darriba D, Taboada GL, Doallo R, Posada D (2012). jModelTest 2: more models, new heuristics and parallel computing.Nat Methods 9, 772-772. |
| 11 | Ebersberger I, Strauss S, von Haeseler A (2009). Ha- MStR: profile hidden markov model based search for orthologs in ESTs.BMC Evol Biol 9, 157. |
| 12 | Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Res 32, 1792-1797. |
| 13 | Feliner GN, Rosselló JA (2007). Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants.Mol Phylogenet Evol 44, 911-919. |
| 14 | Funk VA, Susanna A, Stuessy TF, Robinson H (2009). Classification of compositae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 171-192. |
| 15 | Goertzen LR, Cannone JJ, Gutell RR, Jansen RK (2003). ITS secondary structure derived from comparative ana- lysis: implications for sequence alignment and phylogeny of the Asteraceae.Mol Phylogenet Evol 29, 216-234. |
| 16 | Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Regev A (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome.Nat Biotechnol 29, 644-652. |
| 17 | Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens- Mack J, Müller KF, Boore JL (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns.Proc Natl Acad Sci USA 104, 19369-19374. |
| 18 | Jansen RK, Holsinger KE, Michaels HJ, Palmer JD (1990). Phylogenetic analysis of chloroplast DNA restric- tion site data at higher taxonomic levels: an example from the Asteraceae.Evolution 44, 2089-2105. |
| 19 | Jansen RK, Michaels HJ, Palmer JD (1991). Phylogeny and character evolution in the Asteraceae based on chloroplast DNA restriction site mapping.Syst Bot 16, 98-115. |
| 20 | Karis PO (1993). Morphological phylogenetics of the As- teraceae-Asteroideae, with notes on character evolution.Plant Syst Evol 186, 69-93. |
| 21 | Karis PO (1994). Phylogeny of Asteraceae: Asteroideae revisited. In: Hind DJN, Beentje HJ, eds. Compositae: Systematics. Kew: Royal Botanic Gardens Publishers. pp. 41-47. |
| 22 | Karis PO, Ryding O (1994). Tribe heliantheae. In: Bremer KE, ed. Asteraceae, Cladistics & Classification. Portland: Timber Press. pp. 559-624. |
| 23 | Kim KJ, Jansen RK (1995). ndhF sequence evolution and the major clades in the sunflower family.Proc Natl Acad Sci USA 92, 10379-10383. |
| 24 | Kim KJ, Jasen RK, Wallace RS, Michaels HJ, Palmer JD (1992). Phylogenetic implications of rbcL sequence varia- tion in the Asteraceae.Ann Mo Bot Gard 79, 428-445. |
| 25 | Lin ZG, Kong HZ, Nei M, Ma H (2006). Origins and evolu- tion of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer.Proc Natl Acad Sci USA 103, 10328-10333. |
| 26 | Liu JQ, Gao TG, Chen ZD, Lu AM (2002). Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae).Mol Phylogenet Evol 23, 307-325. |
| 27 | Liu L, Wu SY, Yu LL (2015). Coalescent methods for estimating species trees from phylogenomic data.J Syst Evol 53, 380-390. |
| 28 | Lockhart PJ, Penny D (2005). The place of Amborella within the radiation of angiosperms.Trends Plant Sci 10, 201-202. |
| 29 | Mandel JR, Dikow RB, Funk VA (2015). Using phylogenomics to resolve mega-families: an example from Compositae.J Syst Evol 53, 391-402. |
| 30 | Mandel JR, Dikow RB, Funk VA, Masalia RR, Staton SE, Kozik A, Burke JM (2014). A target enrichment method for gathering phylogenetic information from hundreds of loci: an example from the Compositae.Appl Plant Sci 2, 130085. |
| 31 | Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T (2014). ASTRAL: genome-scale coales- cent-based species tree estimation.Bioinformatics 30, 541-548. |
| 32 | Moore MJ, Bell CD, Soltis PS, Soltis DE (2007). Using plastid genome-scale data to resolve enigmatic relation- ships among basal angiosperms.Proc Natl Acad Sci USA 104, 19363-19368. |
| 33 | Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots.Proc Natl Acad Sci USA 107, 4623-4628. |
| 34 | Nesom GL, Robinson H (2007). Astereae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 284-342. |
| 35 | Ness RW, Graham SW, Barrett SC (2011). Reconciling gene and genome duplication events: using multiple nuclear gene families to infer the phylogeny of the aquatic plant family Pontederiaceae.Mol Biol Evol 28, 3009-3018. |
| 36 | Ng M, Yanofsky MF (2001). Function and evolution of the plant MADS-box gene family.Nat Rev Genet 2, 186-195. |
| 37 | Nordenstam B, Källersjö M (2009). Calenduleae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 527-538. |
| 38 | Nordenstam B, Pelser PB, Kadereit JW, Watson LE (2009). Senecioneae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 503-526. |
| 39 | Oberprieler C, Himmelreich S, Källersjö M, Vallès J, Watson LE, Vogt R (2009). Anthemideae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 631-666. |
| 40 | Oberprieler C, Vogt R, Watson LE (2007). Anthemideae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 243-374. |
| 41 | Panero JL (2005). New combinations and infrafamilial taxa in the Asteraceae. Phytologia 87, 1-14. |
| 42 | Panero JL (2007a). Key to the tribes of the Heliantheae alliance. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 391-395. |
| 43 | Panero JL (2007b). Tribe Coreopsideae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 406-417. |
| 44 | Panero JL (2007c). Tribe Millerieae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 477-492. |
| 45 | Panero JL, Freire SE, Espinar LA, Crozier BS, Barboza GE, Cantero JJ (2014). Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae.Mol Phylogenet Evol 80, 43-53. |
| 46 | Panero JL, Funk VA (2002). Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae).P Biol Soc Wash 115, 909-922. |
| 47 | Panero JL, Funk VA (2008). The value of sampling ano- malous taxa in phylogenetic studies: major clades of the Asteraceae revealed.Mol Phylogenet Evol 47, 757-782. |
| 48 | Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit JW, Watson LE (2010). Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies.Am J Bot 97, 856-873. |
| 49 | Pelser PB, Watson L (2009). Introduction to Asteroideae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxanomy. pp. 495-502. |
| 50 | Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Quackenbush J (2003). TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets.Bioinformatics 19, 651-652. |
| 51 | Phillips MJ, Delsuc F, Penny D (2004). Genome-scale phylogeny and the detection of systematic biases.Mol Biol Evol 21, 1455-1458. |
| 52 | Rieseberg LH, Soltis DE (1991). Phylogenetic consequences of cytoplasmic gene flow in plants.Evol Trends Plants 5, 65-84. |
| 53 | Robinson H (1981). A revision of the tribal and subtribal limits of the Heliantheae (Asteraceae).Smithsonian Contrib Bot 51, 1-102. |
| 54 | Robinson H (2004). New supertribes, Helianthodae and Senecionodae, for the subfamily Asteroideae (Asterac- eae).Phytologia 86, 116-120. |
| 55 | Robinson H (2005). Validation of the supertribe Asterodae.Phytologia 87, 73-74. |
| 56 | Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models.Bioinformatics 19, 1572-1574. |
| 57 | Sang T, Zhong Y (2000). Testing hybridization hypotheses based on incongruent gene trees.Syst Biol 49, 422-434. |
| 58 | Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Soltis PS (2009). Polyploidy and angiosperm diversification.Am J Bot 96, 336-348. |
| 59 | Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu YL, Chase MW, Soltis PS (2004). Genome-scale data, angiosperm relationships, and ‘ending incongruence’: a cautionary tale in phylogenetics.Trends Plant Sci 9, 477-483. |
| 60 | Soltis DE, Kuzoff RK (1995). Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxi- fragaceae).Evolution 49, 727-742. |
| 61 | Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.Bioinformatics 22, 2688-2690. |
| 62 | Stefanović S, Rice DW, Palmer JD (2004). Long branch attraction, taxon sampling, and the earliest angiosperms: amborella or monocots?BMC Evol Biol 4, 35. |
| 63 | Stuessy TF (1977). Heliantheae-systematic review. In: Hey- wood VH, Harborne JB, Turner BL, eds. The Biology and Hemistry of the Compositae. London: Academic Press. pp. 621-671. |
| 64 | Suyama M, Torrents D, Bork P (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments.Nucleic Acids Res 34, W609-W612. |
| 65 | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: molecular evolutionary genetics analysis version 6.0.Mol Biol Evol 30, 2725-2729. |
| 66 | Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Leebens-Mack J (2014). Phylotranscrip- tomic analysis of the origin and early diversification of land plants.Proc Natl Acad Sci USA 111, E4859-E4868. |
| 67 | Xu GX, Ma H, Nei M, Kong HZ (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification.Proc Natl Acad Sci USA 106, 835-840. |
| 68 | Yang Y, Moore MJ, Brockington SF, Soltis DE, Wong GKS, Carpenter EJ, Smith SA (2015). Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing.Mol Biol Evol (in press) |
| 69 | Zeng LP, Zhang Q, Sun RR, Kong HZ, Zhang N, Ma H (2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times.Nat Commun 5, 4956. |
| 70 | Zimmer EA, Wen J (2012). Using nuclear gene data for plant phylogenetics: progress and prospects.Mol Phylogenet Evol 65, 774-785. |
| 71 | Zimmer EA, Wen J (2015). Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches.J Syst Evol 53, 371-379. |
/
| 〈 |
|
〉 |