短穗竹居群遗传结构及气候适应性分析
- 1西南林业大学云南生物多样性研究院, 昆明 650224
2中国科学院昆明植物研究国西南野生生物种质资源库, 昆明 650201
收稿日期: 2024-06-19
录用日期: 2024-11-15
网络出版日期: 2024-11-26
基金资助
国家自然科学基金(31100148);云南省高层次人才培养计划青年拔尖人才专项(YNWR-QNBJ-2019-148)
Population Genetic Structure and Climate Adaptation Analysis of Brachystachyum densiflorum
- 1Yunnan Academy of Biodiversity, Southwest Forestry University, Kunming 650224, China
2Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China
Received date: 2024-06-19
Accepted date: 2024-11-15
Online published: 2024-11-26
摘要
本文引用格式
张如礼 , 李德铢 , 张玉霄 . 短穗竹居群遗传结构及气候适应性分析[J]. 植物学报, 2025 , 60(3) : 407 -424 . DOI: 10.11983/CBB24094
Abstract
INTRODUCTION: Genetic diversity is considered as a crucial aspect in assessment and conservation of rare and endangered species. Brachystachyum densiflorum is a species endemic to eastern China. In recent years, with rapid economic development, accelerated urbanization, and escalating pollutant emissions, the habitat of B. densiflorum has been continuously degraded, habitat fragmentation has intensified, and its populations have shown a tendency to decline.
RATIONALE: Genetic diversity endows species with abundant genetic resources and plays a pivotal role in shaping their capacity to adapt to new environments. To elucidate the genetic diversity of B. densiflorum and evaluate the influence of climate change on its genetic variation, reduced-representation genome sequencing technology was employed to obtain single nucleotide polymorphisms (SNPs), and subsequently population genetics and landscape genetics together with species distribution modelling were analyzed.
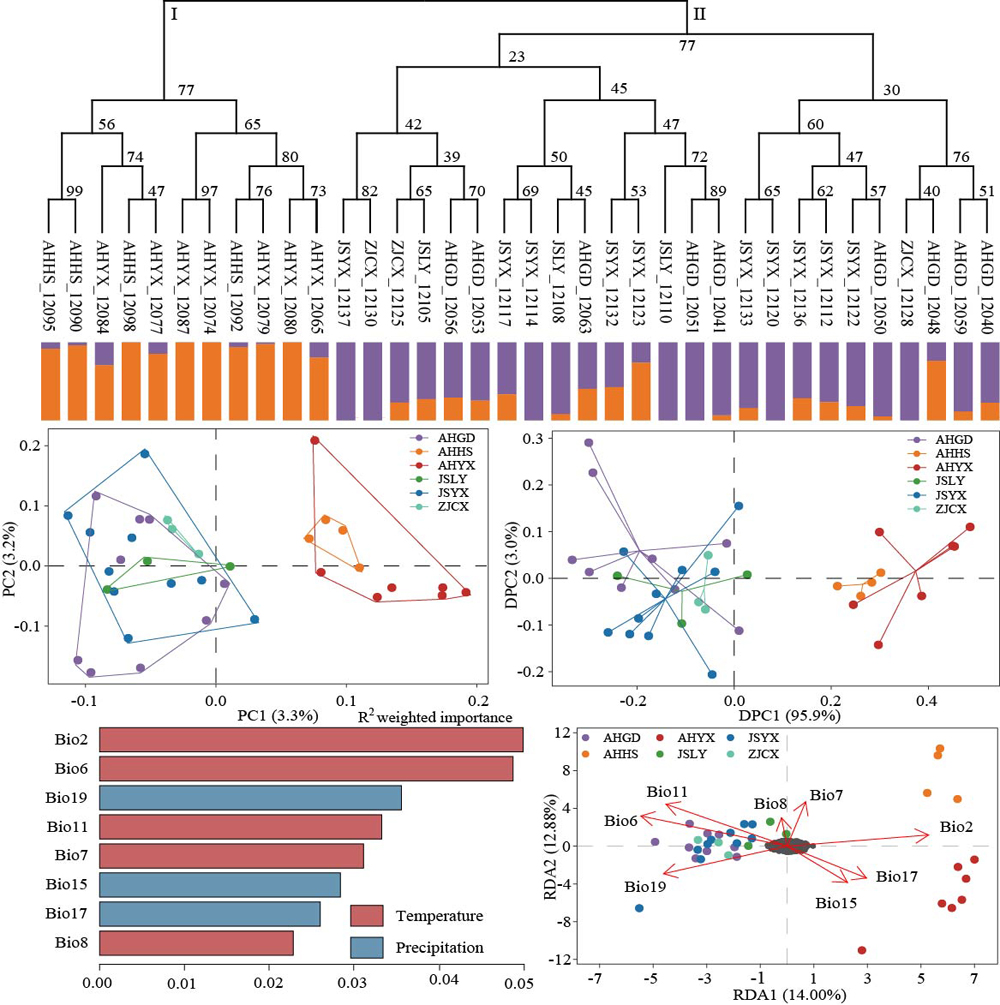
RESULTS: B. densiflorum had a moderate level of genetic diversity. Six populations were divided into two groups, and there was moderate differentiation (FST=0.102) and high gene flow (Nm=2.442) between them. Genotype-environment association analysis indicated that the two groups were diverged attributable to local adaptation to the climate. Temperature differences and low-temperature regimes interacting together with precipitation gave rise to genetic variation of this species. In total, 544 adaptive loci were identified, which displayed significant correlations with temperature difference, low-temperature factors (Bio2, Bio6, Bio11, and Bio7), and precipitation factors (Bio19). B. densiflorum migrated evidently northward from the Last Glacial Maximum to the current, with its distribution area increased by 89.5%. However, during the period from 2061 to 2080, the extent of the suitable area for this species will be contracted, and there will be partial degradation and fragmentation occurring in highly suitable areas within Anhui Province.
CONCLUSION: B. densiflorum showed a moderate level of genetic diversity and a moderate degree of genetic differentiation. Local adaptation drove the formation of the current genetic pattern of B. densiflorum, and temperature differences, low-temperature, and precipitation led to genetic variation. B. densiflorum has evidently migrated northward from the Last Glacial Maximum to the current with increase of distribution area. However, niche modelling indicated that during the period from 2061 to 2080, the suitable habitat area of B. densiflorum would be contracted, with partial degradation and fragmentation occurring in highly suitable areas within Anhui Province. These results provide the basis for conservation and utilization of B. densiflorum.

Population genetic structure analysis of Brachystachyum densiflorum

参考文献
| [1] | Aguirre-Liguori JA, Ramírez-Barahona S, Gaut BS (2021). The evolutionary genomics of species' responses to climate change. Nat Ecol Evol 5, 1350-1360. |
| [2] | Alsos IG, Ehrich D, Thuiller W, Eidesen PB, Tribsch A, Sch?nswetter P, Lagaye C, Taberlet P, Brochmann C (2012). Genetic consequences of climate change for northern plants. Proc Roy Soc B 279, 2042-2051. |
| [3] | Alvarado AH, Bossu CM, Harrigan RJ, Bay RA, Nelson ARP, Smith TB, Ruegg KC (2022). Genotype-environment associations across spatial scales reveal the importance of putative adaptive genetic variation in divergence. Evol Appl 15, 1390-1407. |
| [4] | Attigala L, Gallaher T, Nason J, Clark LG (2017). Genetic diversity and population structure of the threatened temperate woody bamboo Kuruna debilis (Poaceae: Bambusoideae: Arundinarieae) from Sri Lanka based on microsatellite analysis. J Natl Sci Found Sri Lanka 45, 53-65. |
| [5] | Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3, e3376. |
| [6] | Barr K, Beichman AC, Kalhori P, Rajbhandary J, Bay RA, Ruegg K, Smith TB (2021). Persistent panmixia despite extreme habitat loss and population decline in the threatened tricolored blackbird (Agelaius tricolor). Evol Appl 14, 674-684. |
| [7] | Beridze B, S?kiewicz K, Walas ?, Thomas PA, Danelia I, Fazaliyev V, Kvartskhava G, Sós J, Dering M (2023). Biodiversity protection against anthropogenic climate change: conservation prioritization of Castanea sativa in the South Caucasus based on genetic and ecological metrics. Ecol Evol 13, e10068. |
| [8] | Bhandawat A, Sharma V, Singh P, Seth R, Nag A, Kaur J, Sharma RK (2019). Discovery and utilization of EST-SSR marker resource for genetic diversity and population structure analyses of a subtropical bamboo, Dendrocalamus hamiltonii. Biochem Genet 57, 652-672. |
| [9] | Boban S, Maurya S, Jha Z (2022). DNA fingerprinting: an overview on genetic diversity studies in the botanical taxa of Indian Bamboo. Genet Resour Crop Evol 69, 469-498. |
| [10] | Bolte CE, Faske TM, Friedline CJ, Eckert AJ (2022). Divergence amid recurring gene flow: complex demographic histories for two North American pine species (Pinus pungens and P. rigida) fit growing expectations among forest trees. Tree Genet Genomes 18, 35. |
| [11] | Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet P (2007). Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conserv Biol 21, 697-708. |
| [12] | Butler JB, Harrison PA, Vaillancourt RE, Steane DA, Tibbits JFG, Potts BM (2022). Climate adaptation, drought susceptibility, and genomic-informed predictions of future climate refugia for the Australian forest tree Eucalyptus globulus. Forests 13, 575. |
| [13] | Cai CN, Hou QX, Ci XQ, Xiao JH, Zhang CY, Li J (2021). Genetic diversity of Horsfieldia hainanensis: an endangered species with extremely small populations. J Trop Subtrop Bot 29, 547-555. (in Chinese) |
| 蔡超男, 侯勤曦, 慈秀芹, 肖建华, 张灿瑜, 李捷 (2021). 极小种群野生植物海南风吹楠的遗传多样性研究. 热带亚热带植物学报 29, 547-555. | |
| [14] | Cheng J, Kao HX, Dong SB (2020). Population genetic structure and gene flow of rare and endangered Tetraena mongolica Maxim. revealed by reduced representation sequencing. BMC Plant Biol 20, 391. |
| [15] | Clark LG (2023). Integrating genomic and morphological data into bamboo taxonomic and evolutionary studies. Plant Divers 45, 123-124. |
| [16] | Das S, Singh YP, Negi YK, Shrivastav PC (2017). Genetic variability in different growth forms of Dendrocalamus strictus: deogun revisited. N Z J For Sci 47, 235. |
| [17] | Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12, 499-510. |
| [18] | DeSaix MG, George TL, Seglund AE, Spellman GM, Zavaleta ES, Ruegg KC (2022). Forecasting climate change response in an alpine specialist songbird reveals the importance of considering novel climate. Divers Distrib 28, 2239-2254. |
| [19] | Dong FS, Lian LS, Sun BW, Chen Z, Zhu YB (2023). Spatial-temporal variation of visibility and its influencing factors in East China. Meteor Sci Technol 51, 510-519. (in Chinese) |
| 董芳淑, 廉丽姝, 孙博雯, 程禛, 朱艳冰 (2023). 华东地区能见度时空变化特征及影响因子分析. 气象科技 51, 510-519. | |
| [20] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [21] | Ely F, Rada F, Fermin G, Clark LG (2019). Ecophysiology and genetic diversity in species of the bamboo Chusquea in the high Andes, Venezuela. Plant Ecol Divers 12, 555-572. |
| [22] | Exposito-Alonso M, Booker TR, Czech L, Gillespie L, Hateley S, Kyriazis CC, Lang PLM, Leventhal L, Nogues-Bravo D, Pagowski V, Ruffley M, Spence JP, Toro Arana SE, Wei? CL, Zess E (2022). Genetic diversity loss in the Anthropocene. Science 3, 1431-1435. |
| [23] | Feng L, Du FK (2022). Landscape genomics in tree conservation under a changing environment. Front Plant Sci 13, 822217. |
| [24] | Fick SE, Hijmans RJ (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37, 4302-4315. |
| [25] | Filipe JC, Rymer PD, Byrne M, Hardy G, Mazanec R, Ahrens CW (2022). Signatures of natural selection in a foundation tree along Mediterranean climatic gradients. Mol Ecol 31, 1735-1752. |
| [26] | Gibson MJS, Moyle LC (2020). Regional differences in the abiotic environment contribute to genomic divergence within a wild tomato species. Mol Ecol 29, 2204-2217. |
| [27] | Guan BC, Gao JJ, Chen W, Gong X, Ge G (2021). The effects of climate change on landscape connectivity and genetic clusters in a small subtropical and warm-temperate tree. Front Plant Sci 12, 671336. |
| [28] | Guo ZH, Ma PF, Yang GQ, Hu JY, Liu YL, Xia EH, Zhong MC, Zhao L, Sun GL, Xu YX, Zhao YJ, Zhang YC, Zhang YX, Zhang XM, Zhou MY, Guo Y, Guo C, Liu JX, Ye XY, Chen YM, Yang Y, Han B, Lin CS, Lu Y, Li DZ (2019). Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol Plant 12, 1353-1365. |
| [29] | Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier MH, Doligez A, Dutech C, Kremer A, Latouche-Hallé C, Troispoux V, Veron V, Degen B (2006). Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Mol Ecol 15, 559-571. |
| [30] | Harrisson KA, Amish SJ, Pavlova A, Narum SR, Telonis- Scott M, Rourke ML, Lyon J, Tonkin Z, Gilligan DM, Ingram BA, Lintermans M, Gan HM, Austin CM, Luikart G, Sunnucks P (2017). Signatures of polygenic adaptation associated with climate across the range of a threatened fish species with high genetic connectivity. Mol Ecol 26, 6253-6269. |
| [31] | Haupt M, Schmid K (2022). Using landscape genomics to infer genomic regions involved in environmental adaptation of soybean genebank accessions. bioRxiv doi: 10.1101/2022.02.18.480989. |
| [32] | He HL, Zheng XF, Wang YQ, Wang WQ, Li MB, Wang SG, Wang J, Wang CM, Zhan H (2022). Effects of climate change and environmental factors on bamboo (Ferrocalamus strictus), a PSESP unique to China. Forests 13, 2108. |
| [33] | Hu W, Zhang ZY, Chen LD, Peng YS, Wang X (2020). Changes in potential geographical distribution of Tsoongiodendron odorum since the Last Glacial Maximum. Chin J Plant Ecol 44, 44-55. (in Chinese) |
| 胡菀, 张志勇, 陈陆丹, 彭焱松, 汪旭 (2020). 末次盛冰期以来观光木的潜在地理分布变迁. 植物生态学报 44, 44-55. | |
| [34] | I?ik K (2011). Rare and endemic species: why are they prone to extinction? Turk J Bot 35, 411-417. |
| [35] | Jia KH, Zhao W, Maier PA, Hu XG, Jin YQ, Zhou SS, Jiao SQ, El-Kassaby YA, Wang TL, Wang XR, Mao JF (2020). Landscape genomics predicts climate change- related genetic offset for the widespread Platycladus orientalis (Cupressaceae). Evol Appl 13, 665-676. |
| [36] | Jiang WX, Bai TD, Dai HM, Wei Q, Zhang WJ, Ding YL (2017). Microsatellite markers revealed moderate genetic diversity and population differentiation of moso bamboo (Phyllostachys edulis)—a primarily asexual reproduction species in China. Tree Genet Genomes 13, 130. |
| [37] | Jing HY, Xiong XS, Jiang F, Pu XC, Ma WH, Li DJ, Liu ZL, Wang ZH (2024). Climate change filtered out resource- acquisitive plants in a temperate grassland in Inner Mongolia, China. Sci China Life Sci 67, 403-413. |
| [38] | Keenan RJ (2015). Climate change impacts and adaptation in forest management: a review. Ann For Sci 72, 145-167. |
| [39] | Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA (2006). Bambuseae (poaceae). In: WuZY, RavenPH, HongDY, Floraof China,eds. Vol. 22. Beijing: Science Press. pp. 7-180. |
| [40] | Li LB, Guo XJ, Peng ZH, Liu GS, Yuan HS, Zhu BC, Yang K (2008). Effect of the quantity of AFLP primer combinations on accurately identifying bamboo genetic relationships. Chin Bull Bot 25, 449-454. (in Chinese) |
| 李潞滨, 郭晓军, 彭镇华, 刘贯水, 袁洪水, 朱宝成, 杨凯 (2008). AFLP引物组合数量对准确研究竹子系统关系的影响. 植物学通报 25, 449-454. | |
| [41] | Li ST, Ramakrishnan M, Vinod KK, Kalendar R, Yrj?l? K, Zhou MB (2020). Development and deployment of high- throughput retrotransposon-based markers reveal genetic diversity and population structure of Asian bamboo. Forests 11, 31. |
| [42] | Lin N, Landis JB, Sun YX, Huang XH, Zhang X, Liu Q, Zhang HJ, Sun H, Wang HC, Deng T (2021). Demographic history and local adaptation of Myripnois dioica (Asteraceae) provide insight on plant evolution in northern China flora. Ecol Evol 11, 8000-8013. |
| [43] | Liu YH, Wang HC, Yang J, Dao ZL, Sun WB (2024). Conservation genetics and potential geographic distribution modeling of Corybas taliensis, a small ‘sky Island' orchid species in China. BMC Plant Biol 24, 11. |
| [44] | Meena RK, Bhandhari MS, Barhwal S, Ginwal HS (2019). Genetic diversity and structure of Dendrocalamus hamiltonii natural metapopulation: a commercially important bamboo species of northeast Himalayas. 3 Biotech 9, 60. |
| [45] | Meena RK, Negi N, Shankhwar R, Bhandari MS, Kant R, Pandey S, Kumar N, Sharma R, Ginwal HS (2023a). Ecological niche modelling and population genetic analysis of Indian temperate bamboo Drepanostachyum falcatum in the western Himalayas. J Plant Res 136, 483-499. |
| [46] | Meena RK, Negi N, Shankhwar R, Bhandari MS, Sharma R (2023b). Population genetic analysis illustrated a high gene diversity and genetic heterogeneity in Himalayacalamus falconeri: a socio-economically important Indian temperate woody bamboo taxon. J Plant Biochem Biotechnol 32, 438-450. |
| [47] | Morin PA, Luikart G, Wayne RK, The SNP Workshop Group (2004). SNPs in ecology, evolution and conservation. Trends Ecol Evol 19, 208-216. |
| [48] | Nilkanta H, Amom T, Tikendra L, Rahaman H, Nongdam P (2017). ISSR marker based population genetic study of Melocanna baccifera (Roxb.) Kurz: a commercially important bamboo of Manipur, North-East India. Scientifica 2017, 3757238. |
| [49] | Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma R, Hedrick PW (2010). Conservation genetics in transition to conservation genomics. Trends Genet 26, 177-187. |
| [50] | Oumer OA, Dagne K, Feyissa T, Tesfaye K, Durai J, Hyder MZ (2020). Genetic diversity, population structure, and gene flow analysis of lowland bamboo |
| [Oxytenanthera abyssinica (A. Rich.) Munro] in Ethiopia. Ecol Evol 10, 11217-11236.] | |
| [51] | Pauls SU, Nowak C, Bálint M, Pfenninger M (2013). The impact of global climate change on genetic diversity within populations and species. Mol Ecol 22, 925-946. |
| [52] | Perez-Alquicira J, Aguilera-Lopez S, Rico Y, Ruiz-Sanchez E (2021). A population genetics study of three native Mexican woody bamboo species of Guadua (Poaceae: Bambusoideae: Bambuseae: Guaduinae) using nuclear microsatellite markers. Bot Sci 99, 542-559. |
| [53] | Phair NL, Nielsen ES, Von der Heyden S (2021). Applying genomic data to seagrass conservation. Biodivers Conserv 30, 2079-2096. |
| [54] | Polic D, Y?ld?r?m Y, Lee KM, Franzén M, Mutanen M, Vila R, Forsman A (2022). Linking large-scale genetic structure of three Argynnini butterfly species to geography and environment. Mol Ecol 31, 4381-4401. |
| [55] | Poupon V, Chakraborty D, Stejskal J, Konrad H, Schueler S, Lstib?rek M (2021). Accelerating adaptation of forest trees to climate change using individual tree response functions. Front Plant Sci 12, 758221. |
| [56] | Salvado P, Aymerich Boixader P, Parera J, Vila Bonfill A, Martin M, Quélennec C, Lewin JM, Delorme-Hinoux V, Bertrand JAM (2022). Little hope for the polyploid endemic Pyrenean Larkspur (Delphinium montanum): evidences from population genomics and ecological niche modeling. Ecol Evol 12, e8711. |
| [57] | Sang YP, Long ZQ, Dan XM, Feng JJ, Shi TT, Jia CF, Zhang XX, Lai Q, Yang GL, Zhang HY, Xu XT, Liu HH, Jiang YZ, Ingvarsson PK, Liu JQ, Mao KS, Wang J (2022). Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat Commun 13, 6541. |
| [58] | Sepúlveda-Espinoza F, Bertin-Benavides A, Hasbún R, Toro-Nú?ez ó, Varas-Myrik A, Alarcón D, Guillemin ML (2022). The impact of pleistocene glaciations and environmental gradients on the genetic structure of Embothrium coccineum. Ecol Evol 12, e9474. |
| [59] | Shafer ABA, Wolf JBW, Alves PC, Bergstr?m L, Bruford MW, Br?nnstr?m I, Colling G, Dalén L, De Meester L, Ekblom R, Fawcett KD, Fior S, Hajibabaei M, Hill JA, Hoezel AR, H?glund J, Jensen EL, Krause J, Kristensen TN, Krützen M, McKay JK, Norman AJ, Ogden R, ?sterling EM, Ouborg NJ, Piccolo J, Popovi? D, Primmer CR, Reed FA, Roumet M, Salmona J, Schenekar T, Schwartz MK, Segelbacher G, Senn H, Thaulow J, Valtonen M, Veale A, Vergeer P, Vijay N, Vilà C, Weissensteiner M, Wennerstr?m L, Wheat CW, Zieliński P (2015). Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30, 78-87. |
| [60] | Silva SMM, Martins K, Costa FHS, de Campos T, Scherwinski-Pereira JE (2020). Genetic structure and diversity of native Guadua species (Poaceae: Bambusoideae) in natural populations of the Brazilian Amazon rainforest. An Acad Bras Ciênc 92, e20190083. |
| [61] | Sun WY, Shu JP, Gu YF, Morigengaowa, Du XJ, Liu BD, Yan YH (2022). Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides. Biodiv Sci 30, 21508. (in Chinese) |
| 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿 (2022). 基于保护基因组学揭示荷叶铁线蕨的濒危机制. 生物多样性 30, 21508. | |
| [62] | Tian B, Yang HQ, Wong KM, Liu AZ, Ruan ZY (2012). ISSR analysis shows low genetic diversity versus high genetic differentiation for giant bamboo, Dendrocalamus giganteus (Poaceae: Bambusoideae), in China populations. Genet Resour Crop Evol 59, 901-908. |
| [63] | Wang GP, Lai H, Bi S, Guo DL, Zhao XP, Chen XL, Liu S, Liu XG, Su YQ, Yi HD, Li GF (2022a). ddRAD-Seq reveals evolutionary insights into population differentiation and the cryptic phylogeography of Hyporhamphus intermedius in Mainland China. Ecol Evol 12, e9053. |
| [64] | Wang J, Feng C, Jiao TL, Von Wettberg EB, Kang M (2017). Genomic signature of adaptive divergence despite strong nonadaptive forces on edaphic islands: a case study of Primulina juliae. Genome Biol Evol 9, 3495-3508. |
| [65] | Wang L, Liu SF, Yang Y, Meng ZN, Zhuang ZM (2022b). Linked selection, differential introgression and recombination rate variation promote heterogeneous divergence in a pair of yellow croakers. Mol Ecol 31, 5729-5744. |
| [66] | Wang SW, Ge QS, Wang F, Wen XY, Huang JB (2013). Abrupt climate changes of Holocene. Chin Geogr Sci 23, 1-12. |
| [67] | Wu RF, Qi JW, Li WB, Wang L, Shen Y, Liu JW, Teng Y, Roos C, Li M (2023). Landscape genomics analysis provides insights into future climate change-driven risk in rhesus macaque. Sci Total Environ 899, 165746. |
| [68] | Yang FM, Cai L, Dao ZL, Sun WB (2022a). Genomic data reveals population genetic and demographic history of Magnolia fistulosa (Magnoliaceae), a plant species with extremely small populations in Yunnan Province, China. Front Plant Sci 13, 811312. |
| [69] | Yang FY, Crossley MS, Schrader L, Dubovskiy IM, Wei SJ, Zhang RZ (2022b). Polygenic adaptation contributes to the invasive success of the Colorado potato beetle. Mol Ecol 31, 5568-5580. |
| [70] | Yang H, Li JL, Milne RI, Tao WJ, Wang Y, Miao JB, Wang WT, Ju T, Tso S, Luo J, Mao KS (2022c). Genomic insights into the genotype-environment mismatch and conservation units of a Qinghai-Tibet Plateau endemic cypress under climate change. Evol Appl 15, 919-933. |
| [71] | Yang HQ, An MY, Gu ZJ, Tian B (2012). Genetic diversity and differentiation of Dendrocalamus membranaceus (Poaceae: Bambusoideae), a declining bamboo species in Yunnan, China, as based on inter-simple sequence repeat (ISSR) analysis. Int J Mol Sci 13, 4446-4457. |
| [72] | Yebeyen D, Nemomissa S, Hailu BT, Zewdie W, Sileshi GW, Rodríguez RL, Woldie TM (2022). Modeling and mapping habitat suitability of highland bamboo under climate change in Ethiopia. Forests 13, 859-875. |
| [73] | Yuan S, Shi Y, Zhou BF, Liang YY, Chen XY, An QQ, Fan YR, Shen Z, Ingvarsson PK, Wang BS (2023). Genomic vulnerability to climate change in Quercus acutissima, a dominant tree species in East Asian deciduous forests. Mol Ecol 32, 1639-1655. |
| [74] | Zhang RL, Liu WY, Zhang YX, Tu DD, Zhu LY, Zhang WJ, Hui CM (2024). Genetic diversity analysis of Melocalamus arrectus based on reduced-representation genome sequencing. Mol Plant Breed 22, 8076-8087. (in Chinese) |
| 张如礼, 刘蔚漪, 张玉霄, 涂丹丹, 朱礼月, 张文君, 辉朝茂 (2024). 基于简化基因组测序的澜沧梨藤竹遗传多样性分析. 分子植物育种 22, 8076-8087. | |
| [75] | Zhang SS, Kang HM, Yang WZ (2019). Population genetic analysis of Nyssa yunnanensis by reduced-representation sequencing technique. Bull Bot Res 39, 899-907. (in Chinese) |
| 张珊珊, 康洪梅, 杨文忠 (2019). 基于简化基因组技术的云南蓝果树群体遗传分析. 植物研究 39, 899-907. | |
| [76] | Zhang X, Sun YX, Landis JB, Zhang JW, Yang LS, Lin N, Zhang HJ, Guo R, Li LJ, Zhang YH, Deng T, Sun H, Wang HC (2020). Genomic insights into adaptation to heterogeneous environments for the ancient relictual Circaeaster agrestis (Circaeasteraceae, Ranunculales). New Phy-tol 228, 285-301. |
| [77] | Zhang YX, Zeng CX, Li DZ (2012). Complex evolution in Arundinarieae (Poaceae: Bambusoideae): incongruence between plastid and nuclear GBSSI gene phylogenies. Mol Phylogenet Evol 63, 777-797. |
| [78] | Zhao HG, Zhang RL, Xiong Y, Ma CL, Zhang YX (2023). Genetic diversity of Cephalostachyum mannii: an endangered species with extremely small populations. Chin Wild Plant Resour 42, 112-120. (in Chinese) |
| 赵虎刚, 张如礼, 熊云, 马长乐, 张玉霄 (2023). 极小种群野生植物独龙江空竹遗传多样性研究. 中国野生植物资源 42, 112-120. | |
| [79] | Zhao HS, Gao ZM, Wang L, Wang JL, Wang SB, Fei BH, Chen CH, Shi CC, Liu XC, Zhang HL, Lou YF, Chen LF, Sun HY, Zhou XQ, Wang SN, Zhang C, Xu H, Li LC, Yang YH, Wei YL, Yang W, Gao Q, Yang HM, Zhao SC, Jiang ZH (2018). Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). GigaScience 7, giy115. |
| [80] | Zhu S, Liu T, Tang Q, Fu L, Tang S (2014). Evaluation of bamboo genetic diversity using morphological and SRAP analyses. Genetika 50, 306-313. |