

植物学报 ›› 2023, Vol. 58 ›› Issue (2): 274-284.DOI: 10.11983/CBB22265 cstr: 32102.14.CBB22265
张凡凡1, 邢新滢1, 石文清2, 沈懿2, 程祝宽2,3,*( )
)
收稿日期:2022-11-20
接受日期:2023-01-10
出版日期:2023-03-01
发布日期:2023-03-15
通讯作者:
*E-mail: 基金资助:
Fanfan Zhang1, Xinying Xing1, Wenqing Shi2, Yi Shen2, Zhukuan Cheng2,3,*( )
)
Received:2022-11-20
Accepted:2023-01-10
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: 摘要: 寡核苷酸荧光原位杂交技术是一种整合了生物信息学分析、DNA高通量合成以及荧光原位杂交实验的新兴技术。该技术适用于任何具有参考基因组的植物物种, 并可根据研究需求设计靶向某个染色体区域、整条染色体或一组染色体的寡核苷酸探针, 目前已成功应用于数种植物的核型分析、染色体变异、种群演化、同源染色体配对、单倍型分析和异源多倍体识别等研究。该文详述了植物寡核苷酸探针的设计、扩增和标记、染色体制备以及荧光原位杂交的具体操作流程, 以期促进寡核苷酸荧光原位杂交技术在细胞遗传学研究中的应用。
张凡凡, 邢新滢, 石文清, 沈懿, 程祝宽. 植物寡核苷酸荧光原位杂交技术方法. 植物学报, 2023, 58(2): 274-284.
Fanfan Zhang, Xinying Xing, Wenqing Shi, Yi Shen, Zhukuan Cheng. Protocals for Oligonucleotide Fluorescence in situ Hybridization in Plants. Chinese Bulletin of Botany, 2023, 58(2): 274-284.
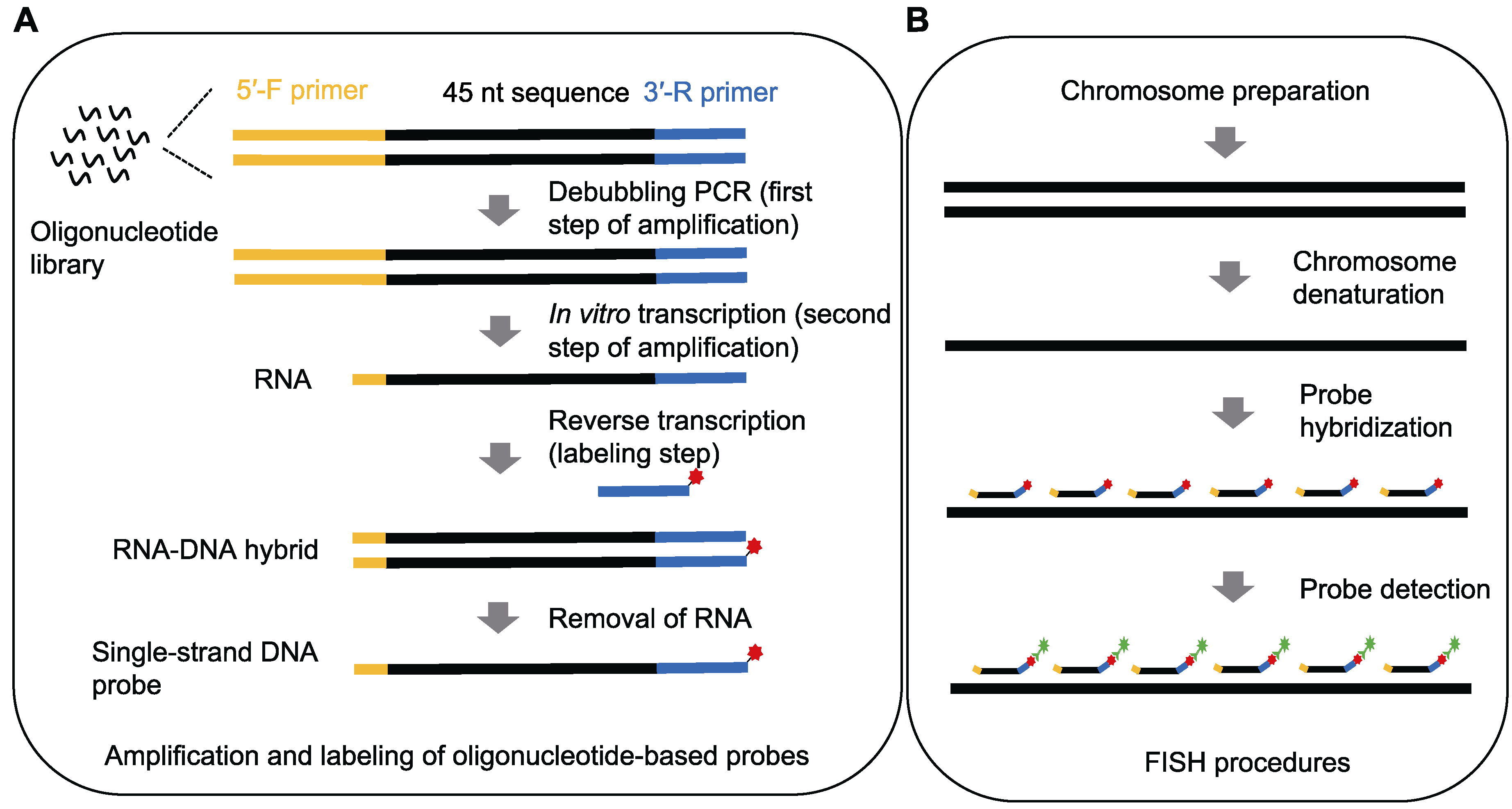
图1 寡核苷酸荧光原位杂交实验流程图 (A) 寡核苷酸探针的扩增和标记; (B) 荧光原位杂交(FISH)实验
Figure 1 Outlines of oligonucleotide fluorescence in situ hybridization (A) Amplification and labeling of oligonucleotide-based probes; (B) Procedures of fluorescence in situ hybridization (FISH)
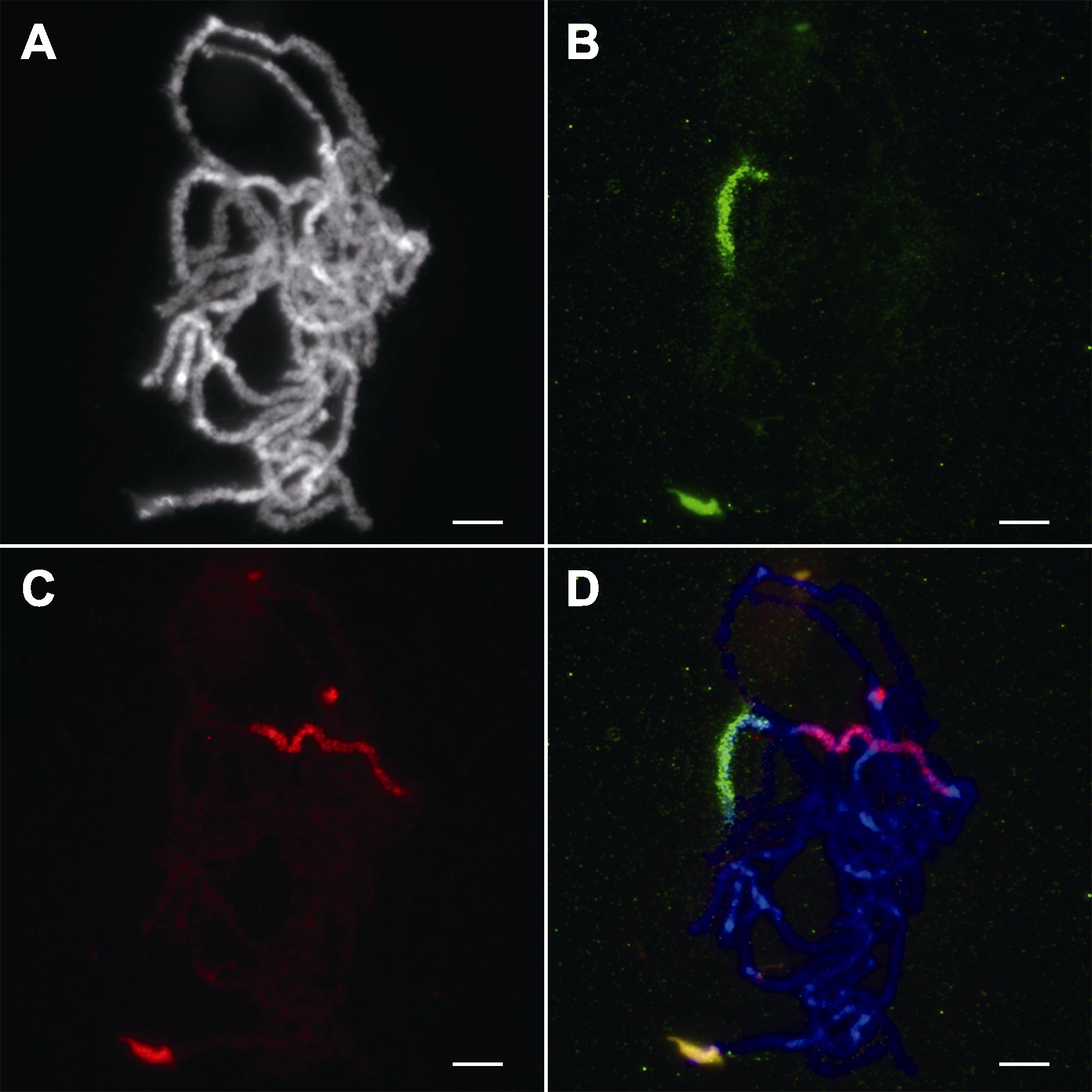
图2 水稻花粉母细胞减数分裂粗线期染色体的寡核苷酸荧光原位杂交鉴定 (A) 粗线期染色体; (B) 特异性靶向水稻11号染色体短臂的寡核苷酸探针(11 S)信号; (C) 特异性靶向水稻11号染色体长臂的寡核苷酸探针(11 L)信号; (D) 三通道合并图(蓝色为染色体, 绿色为11 S, 红色为11 L)。Bars=5 μm
Figure 2 Characterization of rice meiotic pachytene chromosomes by oligonucleotide fluorescence in situ hybridization (A) Pachytene chromosomes; (B) The signal of oligonucleotide probe specific to the short arm of chromosome 11 in rice (11 S); (C) The signal of oligonucleotide probe specific to the long arm of chromosome 11 in rice (11 L); (D) Three-channel combined panel (Blue: Chromosomes; Green: 11 S; Red: 11 L). Bars=5 μm
| [1] |
程新杰, 于恒秀, 程祝宽 (2019). 水稻减数分裂染色体分析方法. 植物学报 54, 503-508.
DOI |
| [2] |
Albert PS, Zhang T, Semrau K, Rouillard JM, Kao YH, Wang CJR, Danilova TV, Jiang JM, Birchler JA (2019). Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc Natl Acad Sci USA 116, 1679-1685.
DOI PMID |
| [3] |
Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang YM, Li JB, Senaratne TN, Williams BR, Rouillard JM, Wu CT (2012). Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA 109, 21301-21306.
DOI PMID |
| [4] |
Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA (2011). Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res 19, 901-909.
DOI URL |
| [5] |
Braz GT, do Vale Martins L, Zhang T, Albert PS, Birchler JA, Jiang JM (2020). A universal chromosome identification system for maize and wild Zea species. Chromosome Res 28, 183-194.
DOI |
| [6] |
Braz GT, He L, Zhao HN, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018). Comparative Oligo- FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208, 513-523.
DOI URL |
| [7] |
Cheng ZK, Buell CR, Wing RA, Jiang JM (2002). Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res 10, 379-387.
DOI URL |
| [8] |
Do Vale Martins L, de Oliveira Bustamante F, da Silva Oliveira AR, da Costa AF, de Lima Feitoza L, Liang QH, Zhao HN, Benko-Iseppon AM, Muñoz-Amatriaín M, Pedrosa-Harand A, Jiang JM, Brasileiro-Vidal AC(2021). BAC- and Oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 130, 133-147.
DOI PMID |
| [9] |
Do Vale Martins L, Yu F, Zhao HN, Dennison T, Lauter N, Wang HY, Deng ZH, Thompson A, Semrau K, Rouillard JM, Birchler JA, Jiang JM (2019). Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10, 4604.
DOI PMID |
| [10] |
Han YH, Zhang T, Thammapichai P, Weng YQ, Jiang JM (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200, 771-779.
DOI URL |
| [11] |
He L, Braz GT, Torres GA, Jiang JM (2018). Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 127, 505-513.
DOI |
| [12] |
Hou LL, Xu M, Zhang T, Xu ZH, Wang WY, Zhang JX, Yu MM, Ji W, Zhu CW, Gong ZY, Gu MH, Jiang JM, Yu HX (2018). Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biol 18, 110.
DOI PMID |
| [13] |
Idziak D, Betekhtin A, Wolny E, Lesniewska K, Wright J, Febrer M, Bevan MW, Jenkins G, Hasterok R (2011). Painting the chromosomes of Brachypodium: current status and future prospects. Chromosoma 120, 469-479.
DOI URL |
| [14] |
Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res 27, 153-165.
DOI |
| [15] |
Jiang JM, Gill BS (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057-1068.
DOI URL |
| [16] |
Liu GQ, Zhang T (2021). Single copy oligonucleotide fluorescence in situ hybridization probe design platforms: development, application and evaluation. Int J Mol Sci 22, 7124.
DOI URL |
| [17] |
Liu XY, Sun S, Wu Y, Zhou Y, Gu SW, Yu HX, Yi CD, Gu MH, Jiang JM, Liu B, Zhang T, Gong ZY (2020). Dual-color Oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101, 112-121.
DOI URL |
| [18] |
Lou QF, Zhang YX, He YH, Li J, Jia L, Cheng CY, Guan W, Yang SQ, Chen JF (2014). Single-copy gene-based chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. Plant J 78, 169-179.
DOI URL |
| [19] |
Lysak MA, Fransz PF, Ali HBM, Schubert I (2001). Chromosome painting in Arabidopsis thaliana. Plant J 28, 689-697.
DOI PMID |
| [20] |
Meng Z, Zhang ZL, Yan TY, Lin QF, Wang Y, Huang WY, Huang YJ, Li ZJ, Yu QY, Wang JP, Wang K (2018). Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a comp- lete set of chromosome-specific Oligo probes. Front Plant Sci 9, 1624.
DOI URL |
| [21] |
Qu MM, Li KP, Han YL, Chen L, Li ZY, Han YH (2017). Integrated karyotyping of woodland strawberry (Fragaria vesca) with Oligopaint FISH probes. Cytogenet Genome Res 153, 158-164.
DOI PMID |
| [22] |
Shi PY, Sun HJ, Liu GQ, Zhang X, Zhou JW, Song RR, Xiao J, Yuan CX, Sun L, Wang ZK, Lou QF, Jiang JM, Wang XE, Wang HY (2022). Chromosome painting reveals inter-chromosomal rearrangements and evolution of subgenome D of wheat. Plant J 112, 55-67.
DOI URL |
| [23] |
Xin HY, Zhang T, Han YH, Wu YF, Shi JS, Xi ML, Jiang JM (2018). Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 127, 313-321.
DOI |
| [24] |
Yamada NA, Rector LS, Tsang P, Carr E, Scheffer A, Sederberg MC, Aston ME, Ach RA, Tsalenko A, Sampas N, Peter B, Bruhn L, Brothman AR (2011). Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet Genome Res 132, 248-254.
DOI PMID |
| [25] |
Yu F, Zhao XW, Chai J, Ding XE, Li XT, Huang YJ, Wang XH, Wu JY, Zhang MQ, Yang QH, Deng ZH, Jiang JM (2022). Chromosome-specific painting unveils chromosomal fusions and distinct allopolyploid species in the Saccharum complex. New Phytol 233, 1953-1965.
DOI URL |
| [26] |
Zhang FF, Shen Y, Miao CB, Cao YW, Shi WQ, Du GJ, Tang D, Li YF, Luo Q, Cheng ZK (2020). OsRAD51D promotes homologous pairing and recombination by preventing nonhomologous interactions in rice meiosis. New Phytol 227, 824-839.
DOI PMID |
| [27] |
Zhang FF, Tang D, Shen Y, Xue ZH, Shi WQ, Ren LJ, Du GJ, Li YF, Cheng ZK (2017). The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell 29, 2597-2609.
DOI URL |
| [28] |
Zhang T, Liu GQ, Zhao HN, Braz GT, Jiang JM (2021). Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J 19, 1967-1978.
DOI PMID |
| [1] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [4] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [5] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [14] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| [15] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||