

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (5): 590-597.DOI: 10.11983/CBB16137
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Received:2016-06-21
Accepted:2017-01-10
Online:2017-09-01
Published:2017-07-10
Contact:
Ying Bao
Ying Bao, Yuqin Mei. Evolution of Defender Against Apoptotic Death (DAD) Genes in Seed Plants[J]. Chinese Bulletin of Botany, 2017, 52(5): 590-597.
| Taxa | Gene ID | Strand | Chromosome | Duplication pattern | Gene structure | Amino acid (aa) | |
|---|---|---|---|---|---|---|---|
| Intron | Exon | ||||||
| Angiosperm | |||||||
| Dicot | |||||||
| Arabidopsis lyrata | AL1G33450 | _ | Scaffold_1 | Block | 4 | 5 | 115 |
| AL4G20890 | + | Scaffold_4 | Block | 4 | 5 | 116 | |
| A. thaliana | AT1G32210 | _ | Chr01 | Block | 4 | 5 | 115 |
| AT2G35520 | + | Chr02 | Block | 4 | 5 | 116 | |
| Brassica rapa | BR05G20160 | + | ChrA05 | Block | 4 | 5 | 115 |
| BR09G26700a | + | ChrA09 | Block | 6 | 7 | 201 (88) d | |
| Capsella rubella | CRU_001G29110 | _ | Scaffold_1 | Block | 4 | 5 | 115 |
| CRU_004G16620 | + | Scaffold_4 | Block | 4 | 5 | 115 | |
| Citrullus lanatus | CL10G00840 | + | Chr10 | 4 | 5 | 115 | |
| Cucumis melo | CM00021G01290 | _ | Scaffold00021 | 4 | 5 | 115 | |
| Eucalyptus grandis | EG0008G05930 | + | Scaffold_8 | 4 | 5 | 115 | |
| Fragaria vesca | FV2G07670 | + | LG2 | 4 | 5 | 124 | |
| Gossypium raimondii | GR03G18540 | + | Chr03 | Block | 4 | 5 | 117 |
| GR08G22400 | _ | Chr08 | Block | 4 | 5 | 117 | |
| Malus domestica | MD05G025840 | + | Chr05 | Random | 4 | 5 | 119 |
| MD10G000120 | + | Chr10 | Random | 4 | 5 | 119 | |
| Manihot esculenta | ME04430G00010 | + | Scaffold04430 | Random | 4 | 5 | 115 |
| ME07304G00010 | + | Scaffold07304 | Random | 4 | 5 | 115 | |
| Populus trichocarpa | PT01G13680 | _ | Chr01 | Block | 4 | 5 | 115 |
| PT03G09680 | + | Chr03 | Block | 4 | 5 | 115 | |
| Prunus persica | PPE_004G33460 | _ | Scaffold_4 | Block | 4 | 5 | 119 |
| PPE_008G00790 | _ | Scaffold_8 | Block | 4 | 5 | 119 | |
| Ricinus communis | RC29634G00390 | + | 29634 | Random | 4 | 5 | 115 |
| RC30068G01440 | + | 30068 | Random | 4 | 5 | 113 | |
| Solanum lycopersicum | SL08G076460 | + | Chr08 | 4 | 5 | 116 | |
| S. tuberosum | ST08G008690 | _ | Chr08 | Block | 4 | 5 | 116 |
| ST08G021840 | _ | Chr08 | Block | 4 | 5 | 116 | |
| Thellungiella parvula | TP1G27890 | _ | Chr1-1 | 4 | 5 | 115 | |
| Theobroma cacao | TC0003G30500 | + | Scaffold_3 | 4 | 5 | 116 | |
| Vitis vinifera | VV02G01690 | + | Chr02 | 4 | 5 | 115 | |
| Moncot | |||||||
| Brachypodium distachyon | BD1G50180 | + | Chr01 | 4 | 5 | 114 | |
| Hordeum vulgare | HV1571041G00020 | _ | Contig_1571041 | Random | 4 | 5 | 114 |
| HV44460G00030 | + | Contig_44460 | Random | 4 | 5 | 114 | |
| Oryza sativa | OS04G32550 | + | Chr04 | 4 | 5 | 114 | |
| Setaria italica | SI004G00880 | + | Scaffold_4 | 4 | 5 | 114 | |
| Sorghum bicolor | SB10G001000 | + | Chr10 | 4 | 5 | 114 | |
| Zea mays | ZM09G06480 | _ | Chr09 | 4 | 5 | 114 | |
| Musa acuminata | MA07G17850 | _ | Chr07 | Block | 4 | 5 | 115 |
| MA10G06540b | _ | Chr10 | Block | - | 1 | 48 (22) d | |
| MA11G08020 | + | Chr11 | Block | 4 | 5 | 170 | |
| Basal taxon | |||||||
| Amborella trichopoda | ATR_00025G00360 | + | Scaffold00025 | 4 | 5 | 121 | |
| Gymnosperm | |||||||
| Taxa | |||||||
| Ginkgo bilobac Gnetum montanumc | Gene ID | Strand | Chromosome | Duplication pattern | Gene structure | Amino acid (aa) | |
| Intron | Exon | ||||||
| Picea abies | GBI00024938 | + | 6973 | - | - | 123 | |
| GMO00028447 | _ | GTHK-0066600 | - | - | 113 | ||
| PAB00021420a | _ | MA_128105 | Random | 5 | 6 | 162 (67) d | |
| P. glaucac | PAB00041636 | _ | MA_42912 | Random | 4 | 5 | 115 |
| PAB00057214b | + | MA_8328929 | Random | 1 | 2 | 50 (45) d | |
| PGL00020840 | + | PUT-39823 PUT-39823 | - | - | 115 | ||
| P. sitchensisc | PGL00013662 | + | PUT-23726 PUT-23726 | - | - | 115 | |
| PGL00010558 | + | PUT-16509 | - | - | 115 | ||
| Pinus pinasterc | PSI00016644 | + | PUT-531483 | - | - | 148 | |
| PSI00008404 | + | PUT-21837 | - | - | 115 | ||
| Pi. sylvestrisc | PPI00061658 | _ | Unigene30039 | - | - | 115 | |
| PPI00006515 | _ | Cotig25413 | - | - | 115 | ||
| PSY00006071 | _ | Isotig31028 | - | - | 115 | ||
| Pi. taeda | PSY00026421 | + | Isotig68699 | - | - | 115 | |
| PSY00004335 | + | isotig24244 | - | - | 115 | ||
| PTA00003657 | _ | Scaffold464 | Random | 4 | 5 | 115 | |
| Pseudotsuga menziesii c | PTA00022345b | + | Scaffold214279 | Random | 2 | 3 | 86 (72) d |
| PTA00022344b | + | Scaffold214279 | Random | 1 | 2 | 44 (44) d | |
| Moss Physcomitrella patens | PME00105748 | + | Psme_598296871 | - | - | 146 | |
| PME00105747 | + | Psme_598296869 | - | - | 115 | ||
| Alga Chlamydomonas reinhardtii | PP00045G01180 | + | Scaffold_45 | 4 | 5 | 131 | |
| PP00456G00180 | - | Scaffold_456 | 4 | 5 | 114 | ||
Table 1 Detailed information of DAD homologous genes of 38 plants
| Taxa | Gene ID | Strand | Chromosome | Duplication pattern | Gene structure | Amino acid (aa) | |
|---|---|---|---|---|---|---|---|
| Intron | Exon | ||||||
| Angiosperm | |||||||
| Dicot | |||||||
| Arabidopsis lyrata | AL1G33450 | _ | Scaffold_1 | Block | 4 | 5 | 115 |
| AL4G20890 | + | Scaffold_4 | Block | 4 | 5 | 116 | |
| A. thaliana | AT1G32210 | _ | Chr01 | Block | 4 | 5 | 115 |
| AT2G35520 | + | Chr02 | Block | 4 | 5 | 116 | |
| Brassica rapa | BR05G20160 | + | ChrA05 | Block | 4 | 5 | 115 |
| BR09G26700a | + | ChrA09 | Block | 6 | 7 | 201 (88) d | |
| Capsella rubella | CRU_001G29110 | _ | Scaffold_1 | Block | 4 | 5 | 115 |
| CRU_004G16620 | + | Scaffold_4 | Block | 4 | 5 | 115 | |
| Citrullus lanatus | CL10G00840 | + | Chr10 | 4 | 5 | 115 | |
| Cucumis melo | CM00021G01290 | _ | Scaffold00021 | 4 | 5 | 115 | |
| Eucalyptus grandis | EG0008G05930 | + | Scaffold_8 | 4 | 5 | 115 | |
| Fragaria vesca | FV2G07670 | + | LG2 | 4 | 5 | 124 | |
| Gossypium raimondii | GR03G18540 | + | Chr03 | Block | 4 | 5 | 117 |
| GR08G22400 | _ | Chr08 | Block | 4 | 5 | 117 | |
| Malus domestica | MD05G025840 | + | Chr05 | Random | 4 | 5 | 119 |
| MD10G000120 | + | Chr10 | Random | 4 | 5 | 119 | |
| Manihot esculenta | ME04430G00010 | + | Scaffold04430 | Random | 4 | 5 | 115 |
| ME07304G00010 | + | Scaffold07304 | Random | 4 | 5 | 115 | |
| Populus trichocarpa | PT01G13680 | _ | Chr01 | Block | 4 | 5 | 115 |
| PT03G09680 | + | Chr03 | Block | 4 | 5 | 115 | |
| Prunus persica | PPE_004G33460 | _ | Scaffold_4 | Block | 4 | 5 | 119 |
| PPE_008G00790 | _ | Scaffold_8 | Block | 4 | 5 | 119 | |
| Ricinus communis | RC29634G00390 | + | 29634 | Random | 4 | 5 | 115 |
| RC30068G01440 | + | 30068 | Random | 4 | 5 | 113 | |
| Solanum lycopersicum | SL08G076460 | + | Chr08 | 4 | 5 | 116 | |
| S. tuberosum | ST08G008690 | _ | Chr08 | Block | 4 | 5 | 116 |
| ST08G021840 | _ | Chr08 | Block | 4 | 5 | 116 | |
| Thellungiella parvula | TP1G27890 | _ | Chr1-1 | 4 | 5 | 115 | |
| Theobroma cacao | TC0003G30500 | + | Scaffold_3 | 4 | 5 | 116 | |
| Vitis vinifera | VV02G01690 | + | Chr02 | 4 | 5 | 115 | |
| Moncot | |||||||
| Brachypodium distachyon | BD1G50180 | + | Chr01 | 4 | 5 | 114 | |
| Hordeum vulgare | HV1571041G00020 | _ | Contig_1571041 | Random | 4 | 5 | 114 |
| HV44460G00030 | + | Contig_44460 | Random | 4 | 5 | 114 | |
| Oryza sativa | OS04G32550 | + | Chr04 | 4 | 5 | 114 | |
| Setaria italica | SI004G00880 | + | Scaffold_4 | 4 | 5 | 114 | |
| Sorghum bicolor | SB10G001000 | + | Chr10 | 4 | 5 | 114 | |
| Zea mays | ZM09G06480 | _ | Chr09 | 4 | 5 | 114 | |
| Musa acuminata | MA07G17850 | _ | Chr07 | Block | 4 | 5 | 115 |
| MA10G06540b | _ | Chr10 | Block | - | 1 | 48 (22) d | |
| MA11G08020 | + | Chr11 | Block | 4 | 5 | 170 | |
| Basal taxon | |||||||
| Amborella trichopoda | ATR_00025G00360 | + | Scaffold00025 | 4 | 5 | 121 | |
| Gymnosperm | |||||||
| Taxa | |||||||
| Ginkgo bilobac Gnetum montanumc | Gene ID | Strand | Chromosome | Duplication pattern | Gene structure | Amino acid (aa) | |
| Intron | Exon | ||||||
| Picea abies | GBI00024938 | + | 6973 | - | - | 123 | |
| GMO00028447 | _ | GTHK-0066600 | - | - | 113 | ||
| PAB00021420a | _ | MA_128105 | Random | 5 | 6 | 162 (67) d | |
| P. glaucac | PAB00041636 | _ | MA_42912 | Random | 4 | 5 | 115 |
| PAB00057214b | + | MA_8328929 | Random | 1 | 2 | 50 (45) d | |
| PGL00020840 | + | PUT-39823 PUT-39823 | - | - | 115 | ||
| P. sitchensisc | PGL00013662 | + | PUT-23726 PUT-23726 | - | - | 115 | |
| PGL00010558 | + | PUT-16509 | - | - | 115 | ||
| Pinus pinasterc | PSI00016644 | + | PUT-531483 | - | - | 148 | |
| PSI00008404 | + | PUT-21837 | - | - | 115 | ||
| Pi. sylvestrisc | PPI00061658 | _ | Unigene30039 | - | - | 115 | |
| PPI00006515 | _ | Cotig25413 | - | - | 115 | ||
| PSY00006071 | _ | Isotig31028 | - | - | 115 | ||
| Pi. taeda | PSY00026421 | + | Isotig68699 | - | - | 115 | |
| PSY00004335 | + | isotig24244 | - | - | 115 | ||
| PTA00003657 | _ | Scaffold464 | Random | 4 | 5 | 115 | |
| Pseudotsuga menziesii c | PTA00022345b | + | Scaffold214279 | Random | 2 | 3 | 86 (72) d |
| PTA00022344b | + | Scaffold214279 | Random | 1 | 2 | 44 (44) d | |
| Moss Physcomitrella patens | PME00105748 | + | Psme_598296871 | - | - | 146 | |
| PME00105747 | + | Psme_598296869 | - | - | 115 | ||
| Alga Chlamydomonas reinhardtii | PP00045G01180 | + | Scaffold_45 | 4 | 5 | 131 | |
| PP00456G00180 | - | Scaffold_456 | 4 | 5 | 114 | ||
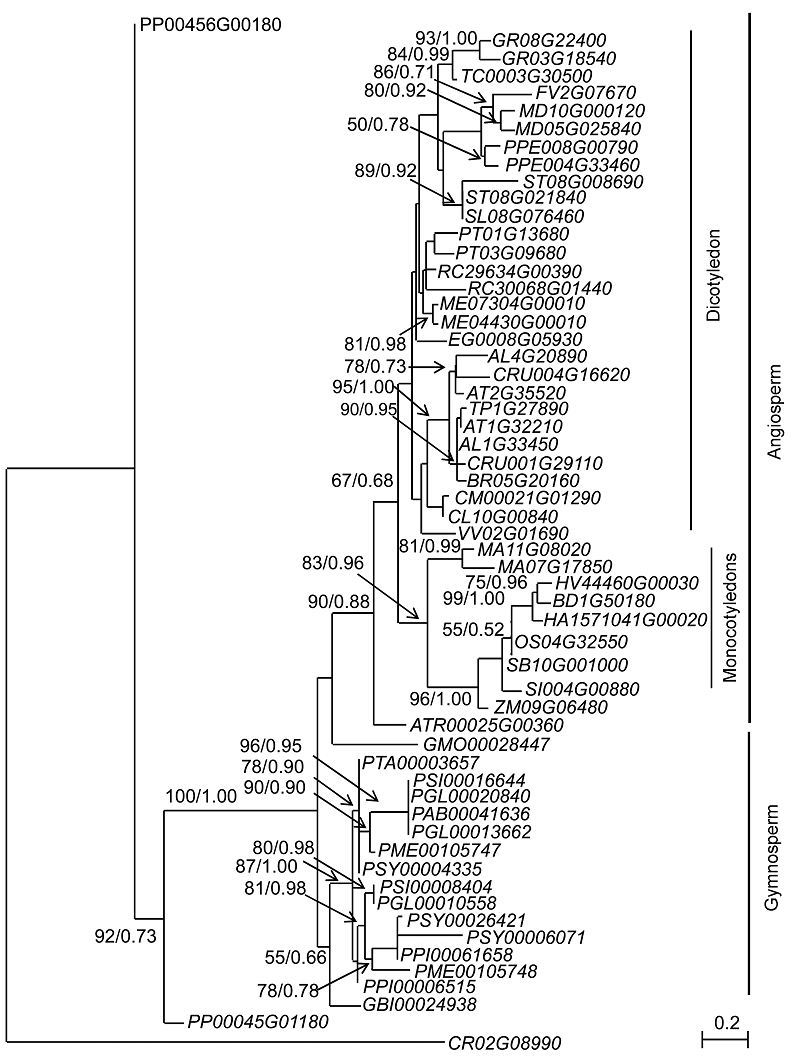
Figure 1 Maximum likelihood tree of DAD genes based on 58 amino acid sequences of 38 plantsNumbers near branches represent bootstrap values (>50%) of the maximum likelihood analysis and posterior rate of the Bayesian analysis. The gene loci in this figure are the same as those listed in Table 1.
| [1] | 龚文芳, 喻树迅, 宋美珍, 范术丽, 庞朝友, 肖水平 (2010). 棉花抗细胞凋亡基因GhDAD1的克隆、定位及表达分析. 中国农业科学 43, 3713-3723. |
| [2] | 贾志蓉, 李丹, 吕应堂 (2004). 拟南芥AtDAD1超量表达植株对H2O2抗性的研究. 武汉植物学研究 22, 373-379. |
| [3] | 杨舒雅, 史娟, 马斌芳, 赵洁, 金晓航 (2012). 抗细胞凋亡因子DAD1的研究进展. 生理科学进展 43, 315-318. |
| [4] | Apte SS, Mattei MG, Seldin MF, Olsen BR (1995). The highly conserved defender against the death 1 (DAD1) gene maps to human chromosome 14q11-q12 and mouse chromosome 14 and has plant and nematode homologs. FEBS Lett 363, 304-306. |
| [5] | Blanc G, Hokamp K, Wolfe KH (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome.Genome Res 13, 137-144. |
| [6] | Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004). Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase- like activities and which can be suppressed by caspase inhibitors, p35 and defender against apoptotic death.J Biol Chem 279, 779-787. |
| [7] | Gallois P, Makishima T, Hecht V, Despres B, Laudié M, Nishimoto T, Cooke R (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant.Plant J 11, 1325-1331. |
| [8] | Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224. |
| [9] | Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.Syst Biol 59, 307-321. |
| [10] | Kelleher DJ, Gilmore R (1997). DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase.Proc Natl Acad Sci USA 94, 4994-4999. |
| [11] | Kerr JFR, Wyllie AH, Currie AR (1972). Apoptosis: a basic biological phenomenon with wide ranging implications in tissue kinetics.Br J Cancer 26, 239-257. |
| [12] | Mondragón-Palomino M, Meyers BC, Michelmore RW, Gaut BS (2002). Patterns of positive selection in the com- plete NBS-LRR gene family of Arabidopsis thaliana.Genome Res 12, 1305-1315. |
| [13] | Nakashima T, Sekiguchi T, Kuraoka A, Fukushima K, Shibata Y, Komiyama S, Nishimoto T (1993). Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells.Mol Cell Biol 13, 6367-6374. |
| [14] | Raff M (1998). Cell suicide for beginners.Nature 396, 119-122. |
| [15] | Roboti P, High S (2012). The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation.J Cell Sci 125, 3474-3484. |
| [16] | Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models.Bioinformatics 19, 1572-1574. |
| [17] | Schwartzman RA, Cidlowski JA (1993). Apoptosis: the biochemistry and molecular biology of programmed cell death.Endocr Rev 14, 133-151. |
| [18] | Williams GT, Smith CA (1993). Molecular regulation of apoptosis: genetic controls on cell death.Cell 7, 777-779. |
| [19] | Wortman JR, Haas BJ, Hannick LI, Smith RK, Maiti R, Ronning CM, Chan AP, Yu CH, Ayele M, Whitelaw CA, White OR, Town CD (2003). Annotation of the Arabidopsis genome.Plant Physiol 132, 461-468. |
| [20] | Ye CY, Li T, Yin H, Weston DJ, Tuskan GA, Tschaplinski TJ, Yang X (2012). Evolutionary analyses of non-family genes in plants.Plant J 73, 788-797. |
| [1] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| [2] | Jia Zhang, Qidong Li, Cui Li, Qinghai Wang, Xincun Hou, Chunqiao Zhao, Shuhe Li, Qiang Guo. Research Progress on MATE Transporters in Plants [J]. Chinese Bulletin of Botany, 2023, 58(3): 461-474. |
| [3] | Jie Tao, Benqiang Li, Jinghua Cheng, Ying Shi, Peihong Liu, Guixia He, Weijie Xu, Huili Liu. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces [J]. Biodiv Sci, 2023, 31(11): 23170-. |
| [4] | Junjie Zhai, Huifeng Zhao, Guangshen Shang, Zhenjun Sun, Yufeng Zhang, Xing Wang. Advances in earthworm genomics: Based on whole genome and mitochondrial genome [J]. Biodiv Sci, 2022, 30(12): 22257-. |
| [5] | Hao Wang, Rui Zhang, Jiao Zhang, Hui Shen, Xiling Dai, Yuehong Yan. De novo transcriptome assembly reveals the whole genome duplication events of Didymochlaena trancatula [J]. Biodiv Sci, 2019, 27(11): 1221-1227. |
| [6] | Shaoshuai Yu, Caili Lin, Shengjie Wang, Wenxin Zhang, Guozhong Tian. Structures of the tuf gene and its upstream part genes and characteristic analysis of conserved regions and activity from related gene promoters of a phytoplasma [J]. Biodiv Sci, 2018, 26(7): 738-748. |
| [7] | Gao Huhu, Zhang Yunxiao, Hu Shengwu, Guo Yuan. Genome-wide Survey and Phylogenetic Analysis of MADS-box Gene Family in Brassica napus [J]. Chinese Bulletin of Botany, 2017, 52(6): 699-712. |
| [8] | Yuanyong Gong, Shuqiao Guo, Hongmei Shu, Wanchao Ni, Paerhati·Maimaiti, Xinlian Shen, Peng Xu, Xianggui Zhang, Qi Guo. Analysis of Molecular Evolution and Gene Structure of EPSPS Protein in Plant Shikimate Pathway [J]. Chinese Bulletin of Botany, 2015, 50(3): 295-309. |
| [9] | Chenlu Xu, Xiaomei Sun, Shougong Zhang. Characteristics of Conifer Genome and Recent Advances in Conifer Sequence Resources Mining [J]. Chinese Bulletin of Botany, 2013, 48(6): 684-693. |
| [10] | Fangzheng Li, Suxin Yang, Chunxia Wu, Haichao Wei, Ruilian Qu, Xianzhong Feng. Structure and Expression Analysis of KNOX Gene Family in Soybean [J]. Chinese Bulletin of Botany, 2012, 47(3): 236-247. |
| [11] | Xiaofeng Dai;Ling Xiao;Yuhua Wu;Gang Wu;Changming Lu. An Overview of Plant Fatty Acid Desaturases and the Coding Genes [J]. Chinese Bulletin of Botany, 2007, 24(01): 105-113. |
| [12] | HUANG Ji HOU Fu-Yun WANG Jian-Fei ZHANG Hong-Sheng. Evolution Styles of Glucose-6-Phosphate Dehydrogenase and 6-Phosphaogluconate Dehydrogenase Genes in Higher Plants [J]. Chinese Bulletin of Botany, 2005, 22(02): 138-146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
