

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (3): 371-377.DOI: 10.11983/CBB18134 cstr: 32102.14.CBB18134
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Quandong Nong1,2,3,Mingyong Zhang1,2,Mei Zhang2,Shuguang Jian2,Hongfang Lu2,Kuaifei Xia1,2,*,Heming Wen3,*
Received:2018-06-11
Accepted:2018-10-14
Online:2019-05-01
Published:2019-11-24
Contact:
Kuaifei Xia,Heming Wen
Quandong Nong,Mingyong Zhang,Mei Zhang,Shuguang Jian,Hongfang Lu,Kuaifei Xia,Heming Wen. Improvement in Extraction Methods of Genomic DNA from Pitaya Stems[J]. Chinese Bulletin of Botany, 2019, 54(3): 371-377.

Figure 1 The washed samples of pitaya mixed with CTAB lysate 1-3: Represented the precipitation can not be mixed with CTAB lysate; 4-9: Represented the precipitation can be mixed with CTAB lysate
| No. | Improved 1 | Improved 2 | Improved 3 | |||
|---|---|---|---|---|---|---|
| OD260/OD280 | OD260/OD230 | OD260/OD280 | OD260/OD230 | OD260/OD280 | OD260/OD230 | |
| 1 | 1.99 | 1.38 | 3.83 | 0.48 | 2.06 | 1.51 |
| 2 | 1.90 | 1.16 | 2.12 | 0.92 | 2.04 | 1.79 |
| 3 | 2.03 | 1.30 | 2.69 | 0.63 | 2.05 | 1.65 |
Table 1 The OD260/OD280 and OD260/OD230 of DNA extracted from pitaya by 3 different improved CTAB+Tris-HCl washing methods
| No. | Improved 1 | Improved 2 | Improved 3 | |||
|---|---|---|---|---|---|---|
| OD260/OD280 | OD260/OD230 | OD260/OD280 | OD260/OD230 | OD260/OD280 | OD260/OD230 | |
| 1 | 1.99 | 1.38 | 3.83 | 0.48 | 2.06 | 1.51 |
| 2 | 1.90 | 1.16 | 2.12 | 0.92 | 2.04 | 1.79 |
| 3 | 2.03 | 1.30 | 2.69 | 0.63 | 2.05 | 1.65 |
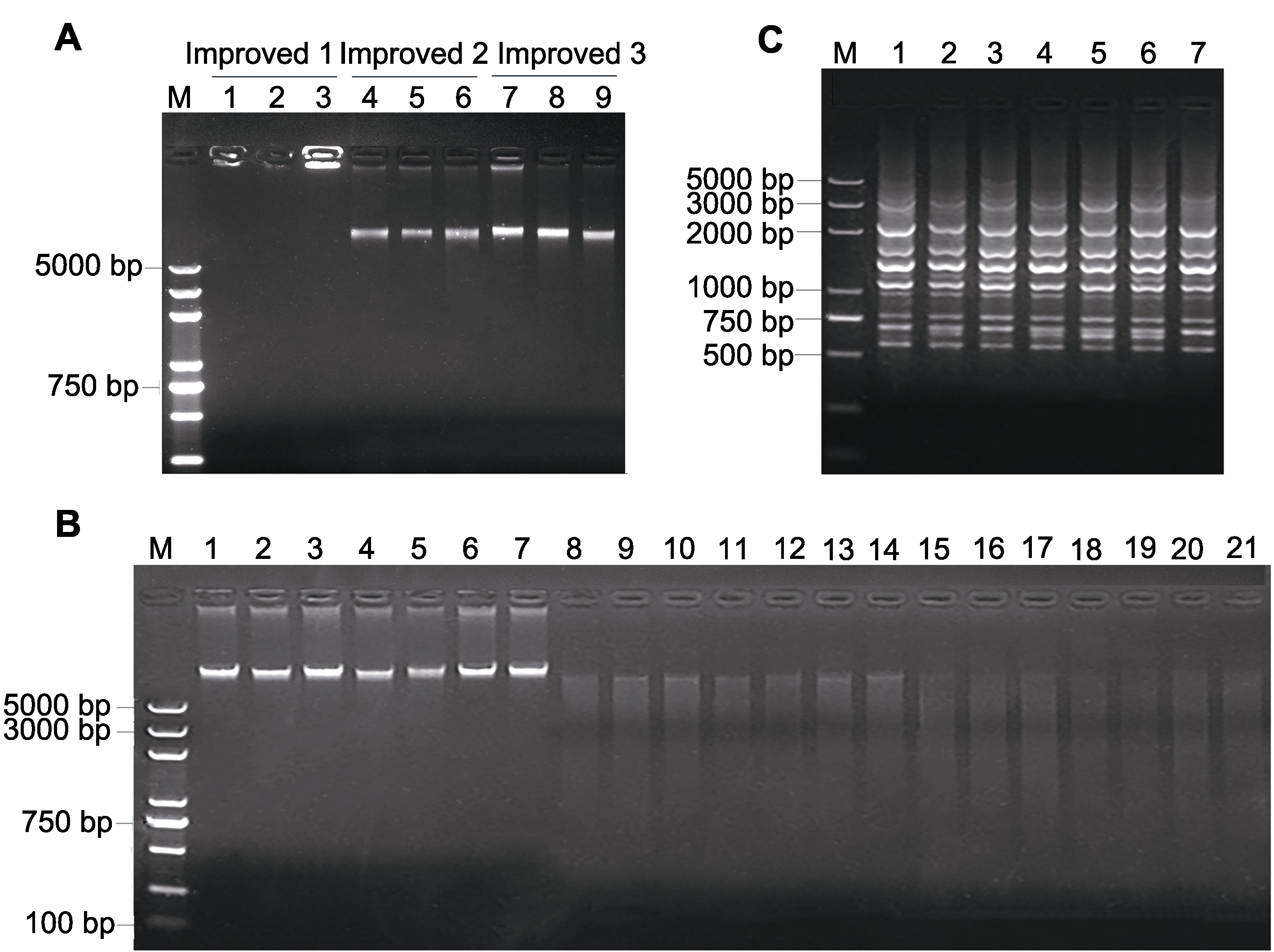
Figure 2 Electrophoretograms of pitaya DNA, enzyme digestion and ISSR amplification (A) Electrophoretogram of DNA extracted by 3 different improved methods (M: DNA marker; Lane 1-3: DNA extracted by improved 1 method; Lane 4-6: DNA extracted by improved 2 method; Lane 7-9: DNA extracted by improved 3 method). (B) Electrophoretogram of pitaya DNA extracted by improved 3 method and enzyme digestion (M: DNA marker; Lane 1-7: DNA; Lane 8-14: DNA digested by HindIII; Lane 15-21: DNA digested by HindIII and EcoRI; The DNA template of 8-14 and 15-21 is corresponding to the DNA in 1-7 lanes, respectively). (C) Electrophoretogram of ISSR amplification (M: DNA marker; Lane 1-7: Products of 7 different DNA templates ISSR amplification).
| [1] | 陈明贤 (2012). 福建省火龙果种质资源ISSR分析及耐盐生理研究. 硕士论文. 福州: 福建农林大学. pp. 11-26. |
| [2] |
程文, 夏正俊, 冯献忠, 杨素欣 (2016). 一种快速、无损大豆种子DNA提取方法的建立和应用. 植物学报 51, 68-73.
DOI URL |
| [3] |
邓仁菊, 范建新, 蔡永强 (2011). 国内外火龙果研究进展及产业发展现状. 贵州农业科学 39(6), 188-192.
DOI URL |
| [4] |
董美超, 岳建强, 李进学, 高俊燕 (2013). 火龙果育种研究进展. 热带农业科学 33(5), 56-59.
DOI URL |
| [5] |
冯建成 (2006). 分子标记辅助选择技术在水稻育种上的应用. 中国农学通报 22(2), 43-47.
DOI URL |
| [6] |
傅建敏, 梁玉琴, 孙鹏, 姬燕晓, 谭晓风 (2015). 柿属植物叶绿体DNA提取方法优化. 中南林业科技大学学报 35(12), 25-28.
DOI URL |
| [7] |
韩静丹, 秦萌, 王献 (2012). 铁线蕨总DNA提取方法比较分析. 河南农业大学学报 46, 182-187.
DOI URL |
| [8] |
李会, 任志莹, 王颖, 王建忠 (2013). 不同DNA提取试剂盒提取作物种子基因组DNA效果的比较. 湖北农业科学 52, 1956-1958.
DOI URL |
| [9] |
李建生 (2007). 玉米分子育种研究进展. 中国农业科技导报 9(2), 10-13.
DOI URL |
| [10] |
李金璐, 王硕, 于婧, 王玲, 周世良 (2013). 一种改良的植物DNA提取方法. 植物学报 48, 72-78.
DOI URL |
| [11] |
梁雪莲, 郑奕雄, 陈晓玲, 曾锦姬 (2007). 花生DNA提取方法比较. 生物技术 17, 41-44.
DOI URL |
| [12] |
罗焜, 马培, 姚辉, 宋经元, 陈科力, 刘义梅 (2012). 中药DNA条形码鉴定中的DNA提取方法研究. 世界科学技术——中医药现代化 14, 1433-1439.
DOI URL |
| [13] | 罗小艳, 郭璇华 (2007). 火龙果的研究现状及发展前景. 食品与发酵工业 33(9), 142-145. |
| [14] | 马小军, 莫长明 (2017). 药用植物分子育种展望. 中国中药杂志 42, 2021-2031. |
| [15] |
邱丽娟, 王昌陵, 周国安, 陈受宜, 常汝镇 (2007). 大豆分子育种研究进展. 中国农业科学 40, 2418-2436.
DOI URL |
| [16] | 申世辉, 马玉华, 蔡永强 (2015). 火龙果研究进展. 中国热带农业 (1), 48-52. |
| [17] | 田新民, 李洪立, 何云, 洪青梅, 胡文斌, 李琼 (2015). 火龙果研究现状. 北方园艺 (18), 188-193. |
| [18] |
吴丽圆 (2004). 4种思茅松总DNA提取方法的比较. 福建林学院学报 24, 237-240.
DOI URL |
| [19] | 吴琳 (2016). 火龙果种质资源评价及繁育研究. 硕士论文. 南宁: 广西大学. pp. 11-61. |
| [20] | 吴敏娜, 武亚琦, 屈艳, 魏纪东, 钟根深 (2015). 四种小鼠肠道微生物DNA提取方法比较. 生态学杂志 34, 1183-1188. |
| [21] | 余志雄, 欧高政, 陈清西, 袁亚芳 (2010). 火龙果总DNA提取方法比较研究. 中国农学通报 26, 300-303. |
| [22] |
詹少华, 尹艺林 (2008). 大豆基因组DNA提取纯化方法研究. 安徽农业科学 36, 9871-9872, 9928.
DOI URL |
| [23] | 张冰雪, 范付华, 乔光, 宋莎, 文晓鹏, 刘涛 (2013). 贵州地方火龙果芽变种质DNA指纹图谱及遗传多样性的ISSR分析. 果树学报 30, 573-577. |
| [24] |
张芬, 李达, 吕长平, 张力, 冯岳东, 许威, 张宏志 (2011). 兰花SRAP指纹图谱的构建. 湖南农业科学 (3), 129-132.
DOI URL |
| [25] |
张桂春, 刘玉静, 李延敏, 牟萍, 曲明娟, 李清, 周菊华 (2017). 火龙果果皮中可溶性膳食纤维的提取方法. 植物学报 52, 622-630.
DOI URL |
| [26] |
张玉晶, 李牡丹, 石旭, 关萍 (2011). 珙桐基因组DNA的提取及ISSR-PCR体系的优化. 山地农业生物学报 30(3), 211-214.
DOI URL |
| [27] |
郑育声, 李东栋, 王哲魁 (2007). 椰子不同组织基因组DNA的提取及质量分析. 海南大学学报自然科学版 25, 51-56.
DOI URL |
| [28] |
Kaczyńska A, Łoś M, Węgrzyn G (2013). An improved method for efficient isolation and purification of genomic DNA from filamentous cyanobacteria belonging to generaAnabaena, Nodularia and Nostoc. Oceanol Hydrobiol Stud 42, 8-13.
DOI URL |
| [29] |
Murray MG, Thompson WF (1980). Rapid isolation of high molecular weight plant DNA.Nucleic Acids Res 8, 4321-4325.
DOI URL |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||