

Chinese Bulletin of Botany ›› 2015, Vol. 50 ›› Issue (6): 683-690.DOI: 10.11983/CBB14147 cstr: 32102.14.CBB14147
Previous Articles Next Articles
Ying Bao*, Jiaxiao Du, Xiang Jing, Si Xu
Received:2014-08-07
Accepted:2015-01-05
Online:2015-11-01
Published:2015-09-06
Contact:
Bao Ying
About author:? These authors contributed equally to this paper
Ying Bao*, Jiaxiao Du, Xiang Jing, Si Xu. Sequence Divergence and Expression Specificity of the Starch Synthase Gene Family in Oryza officinalis Leaf[J]. Chinese Bulletin of Botany, 2015, 50(6): 683-690.
| Gene | SSI | SSII1 | SSII2 | SSII3 | SSIII1 | SSIII2 | SSIV1 | SSIV2 | SSV | GBSSI | GBSSII |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mapped reads | 1 216 | 419 | 3 035 | 82 | 3 365 | 280 | 20 | 249 | 252 | 31 | 9 779 |
| Gene length (bp) | 1 883 | 2 256 | 2 073 | - | 3 663 | 4 653 | - | 2 754 | 2 115 | - | 1 827 |
Table 1 Starch synthase genes in leaf transcriptome of Oryza officinalis
| Gene | SSI | SSII1 | SSII2 | SSII3 | SSIII1 | SSIII2 | SSIV1 | SSIV2 | SSV | GBSSI | GBSSII |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mapped reads | 1 216 | 419 | 3 035 | 82 | 3 365 | 280 | 20 | 249 | 252 | 31 | 9 779 |
| Gene length (bp) | 1 883 | 2 256 | 2 073 | - | 3 663 | 4 653 | - | 2 754 | 2 115 | - | 1 827 |
| Gene | SSI | SSII1 | SSII2 | SSIII1 | SSIII2 | SSIV2 | SSV | GBSSII |
|---|---|---|---|---|---|---|---|---|
| mRNA identities | 96.4% | 95.2% | 95.5% | 97.1% | 96.4% | 95.4% | 95.2% | 98.1% |
| Amino acid identities | 96.9% | 94.5% | 93.1% | 96.3% | 94.1% | 94.7% | 93.6% | 97.0% |
Table 2 Homologous comparisons of the starch synthase genes between Oryza officinalis and O. sativa
| Gene | SSI | SSII1 | SSII2 | SSIII1 | SSIII2 | SSIV2 | SSV | GBSSII |
|---|---|---|---|---|---|---|---|---|
| mRNA identities | 96.4% | 95.2% | 95.5% | 97.1% | 96.4% | 95.4% | 95.2% | 98.1% |
| Amino acid identities | 96.9% | 94.5% | 93.1% | 96.3% | 94.1% | 94.7% | 93.6% | 97.0% |
| SSI | SSII1 | SSII2 | SSIII1 | SSIII2 | SSIV2 | SSV | |
|---|---|---|---|---|---|---|---|
| SSII1 | 41.3% | - | - | - | - | - | - |
| SSII2 | 41.8% | 58.1% | - | - | - | - | - |
| SSIII1 | 28.7% | 23.6% | 25.6% | - | - | - | - |
| SSIII2 | 28.1% | 23.4% | 26.0% | 61.9% | - | - | - |
| SSIV2 | 28.0% | 24.2% | 27.6% | 28.9% | 29.8% | - | - |
| SSV | 20.4% | 18.8% | 20.6% | 24.1% | 24.3% | 27.7% | - |
| GBSSII | 31.6% | 34.4% | 33.5% | 26.9% | 27.1% | 28.6% | 17.8% |
Table 3 Amino acid sequence identities among the starch synthase genes in Oryza officinalis
| SSI | SSII1 | SSII2 | SSIII1 | SSIII2 | SSIV2 | SSV | |
|---|---|---|---|---|---|---|---|
| SSII1 | 41.3% | - | - | - | - | - | - |
| SSII2 | 41.8% | 58.1% | - | - | - | - | - |
| SSIII1 | 28.7% | 23.6% | 25.6% | - | - | - | - |
| SSIII2 | 28.1% | 23.4% | 26.0% | 61.9% | - | - | - |
| SSIV2 | 28.0% | 24.2% | 27.6% | 28.9% | 29.8% | - | - |
| SSV | 20.4% | 18.8% | 20.6% | 24.1% | 24.3% | 27.7% | - |
| GBSSII | 31.6% | 34.4% | 33.5% | 26.9% | 27.1% | 28.6% | 17.8% |
| Sequence | Method | Ka | Ks | Ka/Ks | P-value (Fisher) |
|---|---|---|---|---|---|
| SSI | GY-HKY | 0.009 | 0.079 | 0.112 | 6.75E-14** |
| SSII1 | GY-HKY | 0.026 | 0.114 | 0.225 | 1.46E-14** |
| SSII2 | GY-HKY | 0.035 | 0.062 | 0.560 | 6.95E-03** |
| SSIII1 | GY-HKY | 0.015 | 0.068 | 0.219 | 5.50E-14** |
| SSIII2 | GY-HKY | 0.027 | 0.076 | 0.358 | 2.37E-11** |
| SSIV2 | GY-HKY | 0.029 | 0.100 | 0.289 | 9.13E-12** |
| SSV | GY-HKY | 0.025 | 0.087 | 0.289 | 1.71E-08** |
| GBSSII | GY-HKY | 0.014 | 0.038 | 0.361 | 3.07E-03** |
| SSI | NG | 0.009 | 0.085 | 0.108 | 1.54E-13** |
| SSII1 | NG | 0.024 | 0.128 | 0.189 | 2.83E-16** |
| SSII2 | NG | 0.034 | 0.065 | 0.526 | 5.94E-03** |
| SSIII1 | NG | 0.015 | 0.069 | 0.215 | 3.33E-13** |
| SSIII2 | NG | 0.026 | 0.080 | 0.329 | 5.39E-12** |
| SSIV2 | NG | 0.029 | 0.101 | 0.285 | 6.06E-11** |
| SSV | NG | 0.024 | 0.095 | 0.255 | 2.00E-09** |
| GBSSII | NG | 0.013 | 0.041 | 0.315 | 8.70E-04** |
| SSI | YN | 0.009 | 0.076 | 0.116 | 1.54E-12** |
| SSII1 | YN | 0.026 | 0.110 | 0.233 | 1.18E-12** |
| SSII2 | YN | 0.034 | 0.062 | 0.558 | 1.47E-02** |
| SSIII1 | YN | 0.015 | 0.062 | 0.245 | 1.11E-11** |
| SSIII2 | YN | 0.027 | 0.070 | 0.389 | 4.50E-09** |
| SSIV2 | YN | 0.029 | 0.096 | 0.302 | 2.61E-10** |
| SSV | YN | 0.025 | 0.087 | 0.289 | 5.49E-08** |
| GBSSII | YN | 0.013 | 0.038 | 0.357 | 2.95E-03** |
Table 4 Gene substitutions between Oryza officinalis and O. sativa
| Sequence | Method | Ka | Ks | Ka/Ks | P-value (Fisher) |
|---|---|---|---|---|---|
| SSI | GY-HKY | 0.009 | 0.079 | 0.112 | 6.75E-14** |
| SSII1 | GY-HKY | 0.026 | 0.114 | 0.225 | 1.46E-14** |
| SSII2 | GY-HKY | 0.035 | 0.062 | 0.560 | 6.95E-03** |
| SSIII1 | GY-HKY | 0.015 | 0.068 | 0.219 | 5.50E-14** |
| SSIII2 | GY-HKY | 0.027 | 0.076 | 0.358 | 2.37E-11** |
| SSIV2 | GY-HKY | 0.029 | 0.100 | 0.289 | 9.13E-12** |
| SSV | GY-HKY | 0.025 | 0.087 | 0.289 | 1.71E-08** |
| GBSSII | GY-HKY | 0.014 | 0.038 | 0.361 | 3.07E-03** |
| SSI | NG | 0.009 | 0.085 | 0.108 | 1.54E-13** |
| SSII1 | NG | 0.024 | 0.128 | 0.189 | 2.83E-16** |
| SSII2 | NG | 0.034 | 0.065 | 0.526 | 5.94E-03** |
| SSIII1 | NG | 0.015 | 0.069 | 0.215 | 3.33E-13** |
| SSIII2 | NG | 0.026 | 0.080 | 0.329 | 5.39E-12** |
| SSIV2 | NG | 0.029 | 0.101 | 0.285 | 6.06E-11** |
| SSV | NG | 0.024 | 0.095 | 0.255 | 2.00E-09** |
| GBSSII | NG | 0.013 | 0.041 | 0.315 | 8.70E-04** |
| SSI | YN | 0.009 | 0.076 | 0.116 | 1.54E-12** |
| SSII1 | YN | 0.026 | 0.110 | 0.233 | 1.18E-12** |
| SSII2 | YN | 0.034 | 0.062 | 0.558 | 1.47E-02** |
| SSIII1 | YN | 0.015 | 0.062 | 0.245 | 1.11E-11** |
| SSIII2 | YN | 0.027 | 0.070 | 0.389 | 4.50E-09** |
| SSIV2 | YN | 0.029 | 0.096 | 0.302 | 2.61E-10** |
| SSV | YN | 0.025 | 0.087 | 0.289 | 5.49E-08** |
| GBSSII | YN | 0.013 | 0.038 | 0.357 | 2.95E-03** |
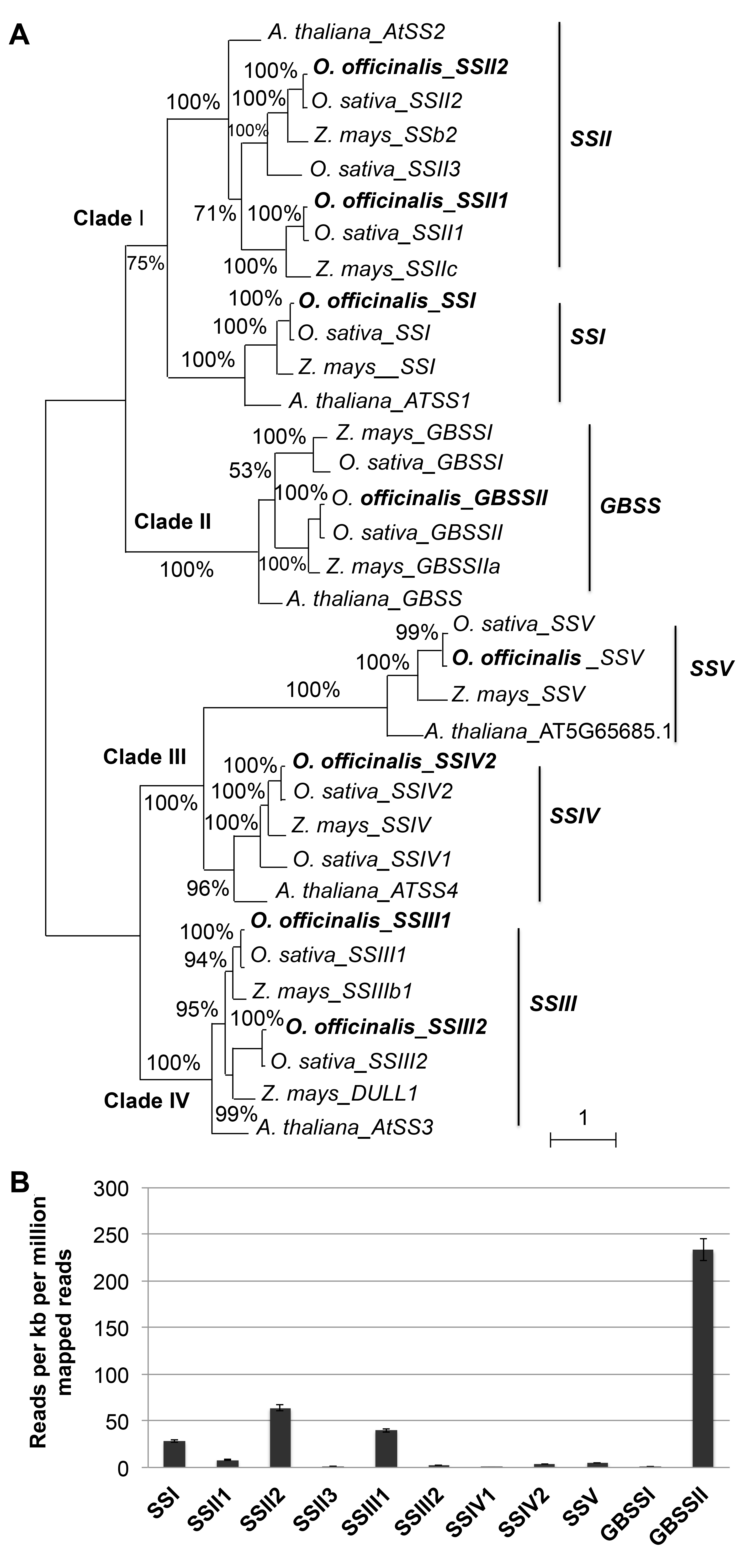
Figure 1 Phylogenetic relationships and relative expression level of the starch synthase gene family(A) The maximum likelihood tree of the starch synthase gene family based on amino acid homologous sequences of Arabidopsis thaliana, Oryza officinalis, O. sativa, and Zea mays (Numbers near the branches indicate bootstrap values); (B) Relative expression level of the starch synthase genes in O. officinalis.
| [1] | Ammiraju JSS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, Talag J, Fan CZ, Goicoechea JL, Zuccolo A, Chen JF, Bennetzen JL, Chen MS, Jackson S, Wing RA (2008). Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set.Plant Cell 20, 3191-3209. |
| [2] | Brust H, Lehmann T, D'Hulst C, Fettke J (2014). Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin.PLoS One 9, e102364. |
| [3] | Buléon A, Colonna P, Planchot V, Ball S (1998). Starch granules: structure and biosynthesis.Int J Biol Macromol 23, 85-112. |
| [4] | Cao HP, Imparl-Radosevich J, Guan HP, Keeling PL, James MG, Myers AM (1999). Identification of the soluble starch synthase activities of maize endosperm.Plant Phy- siol 120, 205-216. |
| [5] | Deschamps P, Moreau H, Worden AZ, Dauvillée D, Ball SG (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts.Genetics 178, 2373-2387. |
| [6] | Dian WM, Jiang HW, Wu P (2005). Evolution and expression analysis of starch synthase III and IV in rice.J Exp Bot 56, 623-632. |
| [7] | Fujita N (2014). Starch biosynthesis in rice endosperm.AGri-Biosci Monogr 4, 1-18. |
| [8] | Fujita N, Satoh R, Hayashi A, Kodama M, Itoh R, Aihara S, Nakamura Y (2011). Starch biosynthesis in rice endosp- erm requires the presence of either starch synthase I or IIIa.J Exp Bot 62, 4819-4831. |
| [9] | Gámez-Arjona FM, Li J, Raynaud S, Baroja-Fernández E, Munñoz FJ, Ovecka M, Ragel P, Bahaji A, Pozueta-Romero J, Mérida Á (2011). Enhancing the expression of starch synthase class IV results in increased levels of both transitory and long-term storage starch.Plant Biotechnol J 9, 1049-1060. |
| [10] | Garber M, Grabherr MG, Guttman M, Trapnell C (2011). Computational methods for transcriptome annotation and quantification using RNA-seq.Nat Methods 8, 469-477. |
| [11] | Goldman N, Yang ZH (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences.Mol Biol Evol 11, 725-736. |
| [12] | Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224. |
| [13] | Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.Syst Biol 59, 307-321. |
| [14] | Hirose T, Terao T (2004). A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.).Planta 220, 9-16. |
| [15] | Jiang HW, Dian WM, Liu FY, Wu P (2004). Molecular cloning and expression analysis of three genes encoding starch synthase II in rice.Planta 218, 1062-1070. |
| [16] | Langmead B, Salzberg SL (2012). Fast gapped-read align- ment with Bowtie 2.Nat Methods 9, 357-359. |
| [17] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools.Bioinformatics 25, 2078-2079. |
| [18] | Lu F, Ammiraju JSS, Sanyal A, Zhang SL, Song RT, Chen JF, Li GS, Sui Y, Song X, Cheng ZK, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen MS (2009). Comparative sequence analysis of MONOCULM1-orthol- ogous regions in 14 Oryza genomes.Proc Natl Acad Sci USA 106, 2071-2076. |
| [19] | Myers AM, Morell MK, James MG, Ball SG (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal.Plant Physiol 122, 989-998. |
| [20] | Nei M, Gojobori T (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions.Mol Biol Evol 3, 418-426. |
| [21] | Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [22] | Orzechowski S (2008). Starch metabolism in leaves.Acta Biochim Pol 55, 435-445. |
| [23] | Schwarte S, Brust H, Steup M, Tiedemann R (2013). Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana.BMC Res Notes 6, 84. |
| [24] | Stamova BS, Laudencia-Chingcuanco D, Beckles DM (2009). Transcriptomic analysis of starch biosynthesis in the developing grain of hexaploid wheat.Int J Plant Genomics 2009, 407426. |
| [25] | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation.Nat Biotechnol 28, 511-515. |
| [26] | Yang ZH, Nielsen R (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models.Mol Biol Evol 17, 32-43. |
| [27] | Zeeman SC, Kossmann J, Smith AM (2010). Starch: its metabolism, evolution, and biotechnological modification in plants.Annu Rev Plant Biol 61, 209-234. |
| [28] | Zeeman SC, Smith SM, Smith AM (2007). The diurnal metabolism of leaf starch.Biochem J 401, 13-28. |
| [29] | Zhang XL, Szydlowski N, Delvallé D, D'Hulst C, James MG, Myers AM (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis.BMC Plant Biol 8, 96. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||