

植物学报 ›› 2019, Vol. 54 ›› Issue (4): 497-502.DOI: 10.11983/CBB19108 cstr: 32102.14.CBB19108
收稿日期:2019-06-13
接受日期:2019-06-20
出版日期:2019-07-01
发布日期:2020-01-08
通讯作者:
左建儒
基金资助:
Lichao Chen,Ni Zhan,Yansha Li,Jian Feng,Jianru Zuo*( )
)
Received:2019-06-13
Accepted:2019-06-20
Online:2019-07-01
Published:2020-01-08
Contact:
Jianru Zuo
摘要: S-亚硝基化是一种重要的蛋白质翻译后修饰方式, 是指一氧化氮(NO)基团共价连接至靶蛋白特定半胱氨酸残基的自由巯基, 从而形成S-亚硝基硫醇(SNO)的过程。S-亚硝基化修饰广泛存在于各有机体中, 通过改变蛋白质生化活性、稳定性、亚细胞定位以及蛋白质-蛋白质相互作用等机制而调控不同的生物学过程或信号通路。在蛋白质S-亚硝基化检测分析方法中, 最为广泛使用的是生物素转化法(biotin switch assay), 其基本原理是首先封闭未被修饰的自由巯基, 进而将被修饰的SNO基团特异地还原为自由巯基并使用生物素将其特异标记。被生物素标记的半胱氨酸残基(即被修饰位点)可进一步通过蛋白质免疫印迹和/或质谱等方法进行检测分析。该文详细描述了植物蛋白质样品的体内和体外生物素转化法的实验流程, 并对实验过程中的注意事项进行了讨论。
陈立超,詹妮,李彦莎,冯健,左建儒. 植物蛋白质S-亚硝基化修饰的检测与分析. 植物学报, 2019, 54(4): 497-502.
Lichao Chen,Ni Zhan,Yansha Li,Jian Feng,Jianru Zuo. Detection and Analysis of Protein S-nitrosylation in Plants. Chinese Bulletin of Botany, 2019, 54(4): 497-502.
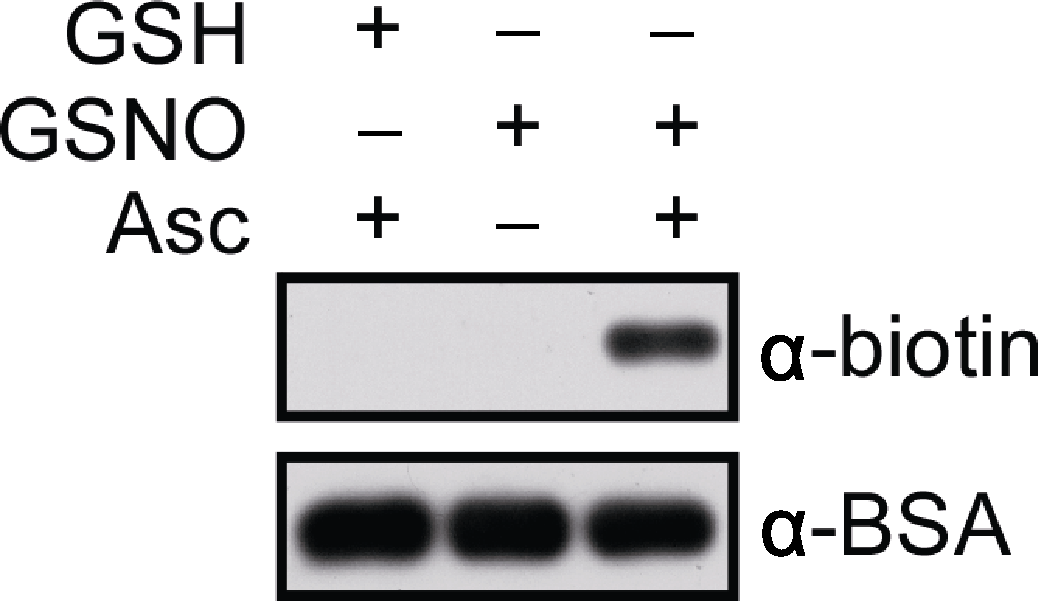
图1 牛血清白蛋白(BSA)体外S-亚硝基化修饰的分析检测 使用biotin-maleimide标记BSA样品, 经SDS-PAGE胶分离后进行免疫印迹分析。分别采用anti-biotin和anti-BSA抗体作为第一抗体(primary antibodies; 1:20 000稀释), anti-mouse IgG为第二抗体(secondary antibodies)。样品信号用SuperSignal Western Femto Maximun Sensitivity Substrate Kit检测。曝光时间分别为20秒(上)和30秒(下)。GSH: 谷胱甘肽; GSNO: S-亚硝基谷胱甘肽; Asc: 抗坏血酸盐
Figure 1 In vitro analysis of S-nitrosylated bovine serum albumin (BSA) BSA is labelled with biotin-maleimide, subjected to SDS- PAGE and western blotting. Anti-biotin and anti-BSA antibodies are used as primary antibodies, respectively (dilution in 1:20 000) and anti-mouse IgG as a secondary antibody. Signals are detected by using the SuperSignal Western Femto Maximun Sensitivity Substrate Kit. The blots are exposed for 20 sec (top) and 30 sec (bottom), respectively. GSH: Glutathione; GSNO: S-nitrosoglutathione; Asc: Sodium ascorbate
| [1] | Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sánchez-Vicente I, Nambara E, Lorenzo O ( 2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun 6, 8669. |
| [2] | Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, Zuo J ( 2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19, 1377. |
| [3] | Cui B, Pan Q, Clarke D, Villarreal MO, Umbreen S, Yuan B, Shan W, Jiang J, Loake GJ ( 2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat Commun 9, 4226. |
| [4] | Feng J, Chen L, Zuo J ( 2019). Protein S-nitrosylation in plants: current progresses and challenges. J Integr Plant Biol doi. org/10.1111/jipb.12780. |
| [5] | Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J ( 2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4, 1529. |
| [6] | He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, Fiorani F, Jackson RB, Crawford NM, Pei ZM ( 2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968-1971. |
| [7] | Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS ( 2005). Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6, 150-166. |
| [8] | Hess DT, Stamler JS ( 2012). Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287, 4411-4418. |
| [9] | Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J ( 2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167, 1731-1746. |
| [10] | Hu J, Yang H, Mu J, Lu T, Peng J, Deng X, Kong Z, Bao S, Cao X, Zuo J ( 2017). Nitric oxide regulates protein methylation during stress responses in plants. Mol Cell 67, 702-710. |
| [11] | Iglesias MJ, Terrile MC, Correa-Aragunde N, Colman SL, Izquierdo-Álvarez A, Fiol DF, París R, Sánchez-López N, Marina A, Calderón Villalobos LIA, Estelle M, Lamattina L, Martínez-Ruiz A, Casalongué CA ( 2018). Regulation of SCFTIR1/AFBs E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol 18, 200-210. |
| [12] | Jaffrey SR, Snyder SH ( 2001). The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001,pl1. |
| [13] | Ling T, Bellin D, Vandelle E, Imanifard Z, Delledonne M ( 2017). Host-mediated S-nitrosylation disarms the bacterial effector HopAI1 to reestablish immunity. Plant Cell 29, 2871-2881. |
| [14] | Lytvyn DI, Raynaud C, Yemets AI, Bergounioux C, Blume YB ( 2016). Involvement of inositol biosynthesis and nitric oxide in the mediation of UV-B induced oxidative stress. Front Plant Sci 7, 430. |
| [15] | Pan QN, Geng CC, Li DD, Xu SW, Mao DD, Umbreen S, Loake GJ, Cui BM ( 2019). Nitrate reductase-mediated nitric oxide regulates the leaf shape in Arabidopsis by mediating the homeostasis of reactive oxygen species. Int J Mol Sci 20, 2235. |
| [16] | París R, Vazquez MM, Graziano M, Terrile MC, Miller ND, Spalding EP, Otegui MS, Casalongué CA ( 2018). Distribution of endogenous NO regulates early gravitropic response and PIN2 localization in Arabidopsis roots. Front Plant Sci 9, 495. |
| [17] | Seth D, Hess DT, Hausladen A, Wang L, Wang YJ, Stamler JS ( 2018). A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol Cell 69, 451-464. |
| [18] | Seth P, Hsieh PN, Jamal S, Wang L, Gygi SP, Jain MK, Coller J, Stamler JS ( 2019). Regulation of microRNA machinery and development by interspecies S-nitrosylation. Cell 176, 1014-1025. |
| [19] | Shekariesfahlan A, Lindermayr C ( 2018). Identification of NO-sensitive cysteine residues using cysteine mutants of recombinant proteins. In: Mengel A, Lindermayr C, eds. Nitric Oxide: Methods and Protocols. New York: Springer New York. pp. 183-203. |
| [20] | Shi H, Liu W, Wei Y, Ye T ( 2017). Integration of auxin/ indole-3-acetic acid 17 and RGA-LIKE3 confers salt stress resistance through stabilization by nitric oxide in Arabidopsis. J Exp Bot 68, 1239-1249. |
| [21] | Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X ( 2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952-956. |
| [22] | Wang P, Zhu JK, Lang Z ( 2015). Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal Behav 10, e1031939. |
| [23] | Willems P, Horne A, Van Parys T, Goormachtig S, De Smet I, Botzki A, Van Breusegem F, Gevaert K ( 2019). The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J 99, 752-762. |
| [24] | Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J ( 2010). GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS One 5, e11290. |
| [25] | Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J ( 2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol 167, 1604. |
| [26] | Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, Liu Z, Zhou Y, Peng J, Wang K, Huang X, Xiao S, Zuo J ( 2018). S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell 71, 142-154. |
| [27] | Zhang L, Shi X, Zhang Y, Wang J, Yang J, Ishida T, Jiang W, Han X, Kang J, Wang X, Pan L, Lv S, Cao B, Zhang Y, Wu J, Han H, Hu Z, Cui L, Sawa S, He J, Wang G ( 2019 a). CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ 42, 1033-1044. |
| [28] | Zhang ZW, Fu YF, Zhou YH, Wang CQ, Lan T, Chen GD, Zeng J, Chen YE, Yuan M, Yuan S, Hu JY ( 2019 b). Nitrogen and nitric oxide regulate Arabidopsis flowering differently. Plant Sci 284, 177-184. |
| [1] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [4] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [5] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [14] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [15] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||